Small RNA Detection by in Situ Hybridization Methods
Abstract
:1. Introduction
2. Approaches for miRNA ISH
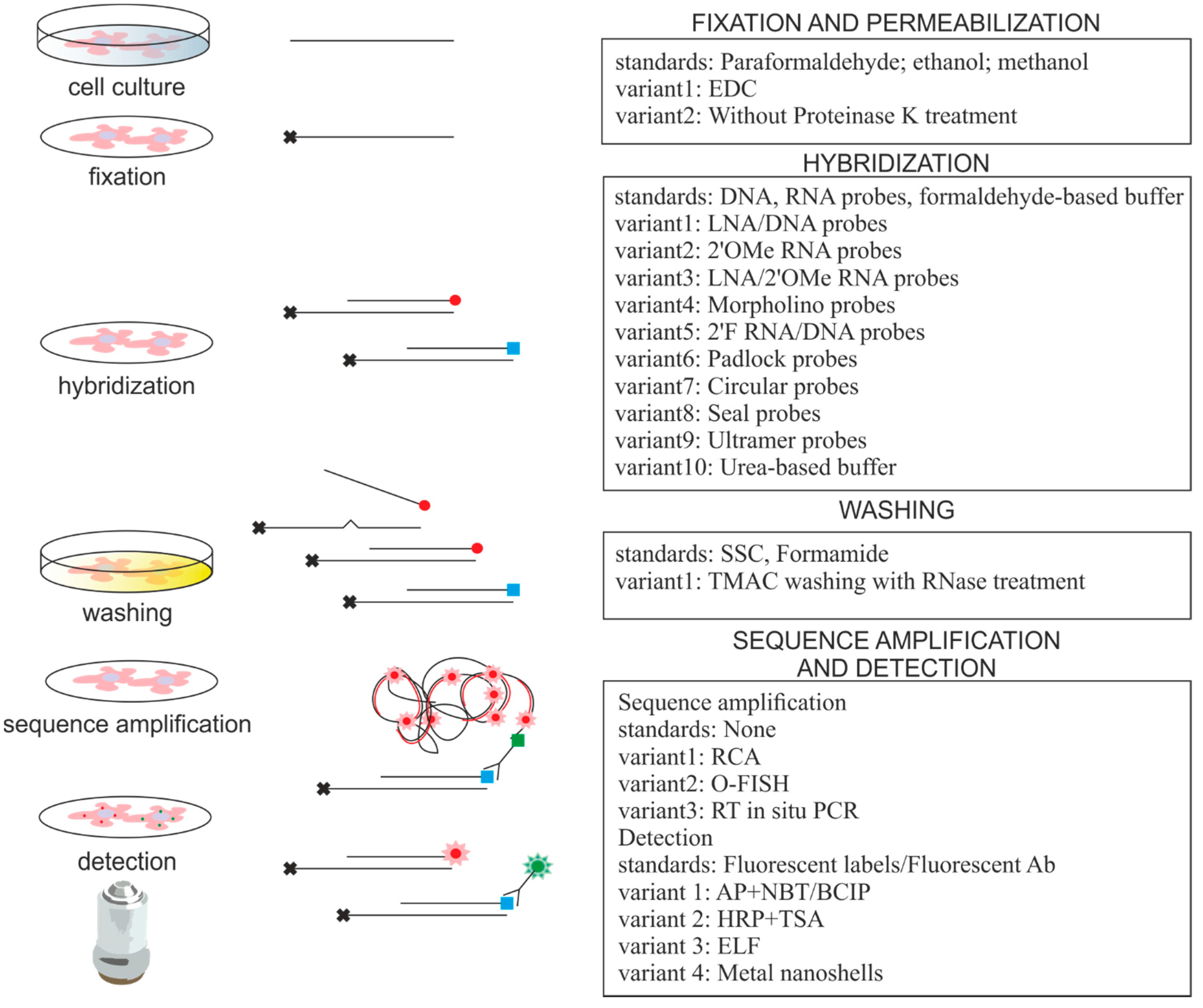
| miRNA ISH Protocol Variations | Advantages | Comments | References |
|---|---|---|---|
| LNA/DNA probes | High specificity and affinity | Golden standard in ISH, expensive | [5,9,19–36] |
| LNA/2′OMe RNA probes | Faster hybridization kinetics and ability to bind structured targets | Probes bind to blocking RNAs | [37,38] |
| RNA probes, TMAC washing, RNase A treatment | Single set of conditions for many probes, Tm for probe-target duplexes independent of GC composition, RNase A treatment decrease off-target binding | Applicable for multiplex analysis | [39,40] |
| 2′F RNA/DNA probes | Increased hybridization efficiency, high selectivity | Applicable for high throughput analysis | [41] |
| Morpholino probes | High specificity and affinity | Hybridization is independent of salt concentration | [42] |
| DNA padlock probes, RCA | Up to single nucleotide specificity, RCA provides signal amplification | Applicable for detection of low abundant miRNA | [43] |
| DNA probes, PLA detection, RCA (O-FISH) | RCA as above | PLA originally used for protein detection | [44] |
| Circular DNA probes, RCA | Fast and efficient protocol, RCA as above | Applicable for multiplex analysis | [45] |
| Seal probes, RCA (TIRCA) | High specificity, decreased loss of miRNAs, RCA as above | Applicable for detection on single molecule level, low protocol temperature | [46] |
| Ultramer probes, RT in situ PCR | Signal amplification | Detects mature miRNAs only | [47] |
| Fluorescent metal nanoshell probes | Improved signal intensity and photostability | Improved optical properties of fluorophores, long lifetime emission signal | [36] |
| EDC fixation | Decreased loss of small RNAs, EDC immobilizes miRNA molecules | Important for low abundant miRNAs detection | [21,22,48,49] |
| NBT/BCIP detection system | Enhanced signal strength | Applicable for detection of low abundant miRNA | [6–8,50–53] |
| TSA detection system | Enhanced signal strength | Applicable for detection of low abundant miRNA | [22,25,31,32,38,48] |
| ELF detection system | High cellular resolution and signal strength | Applicable for detection of low abundant miRNA, single molecule detection, high photostability of precipitate, short exposure time | [41,49] |
2.1. Probes
2.1.1. Directly Labeled Probes
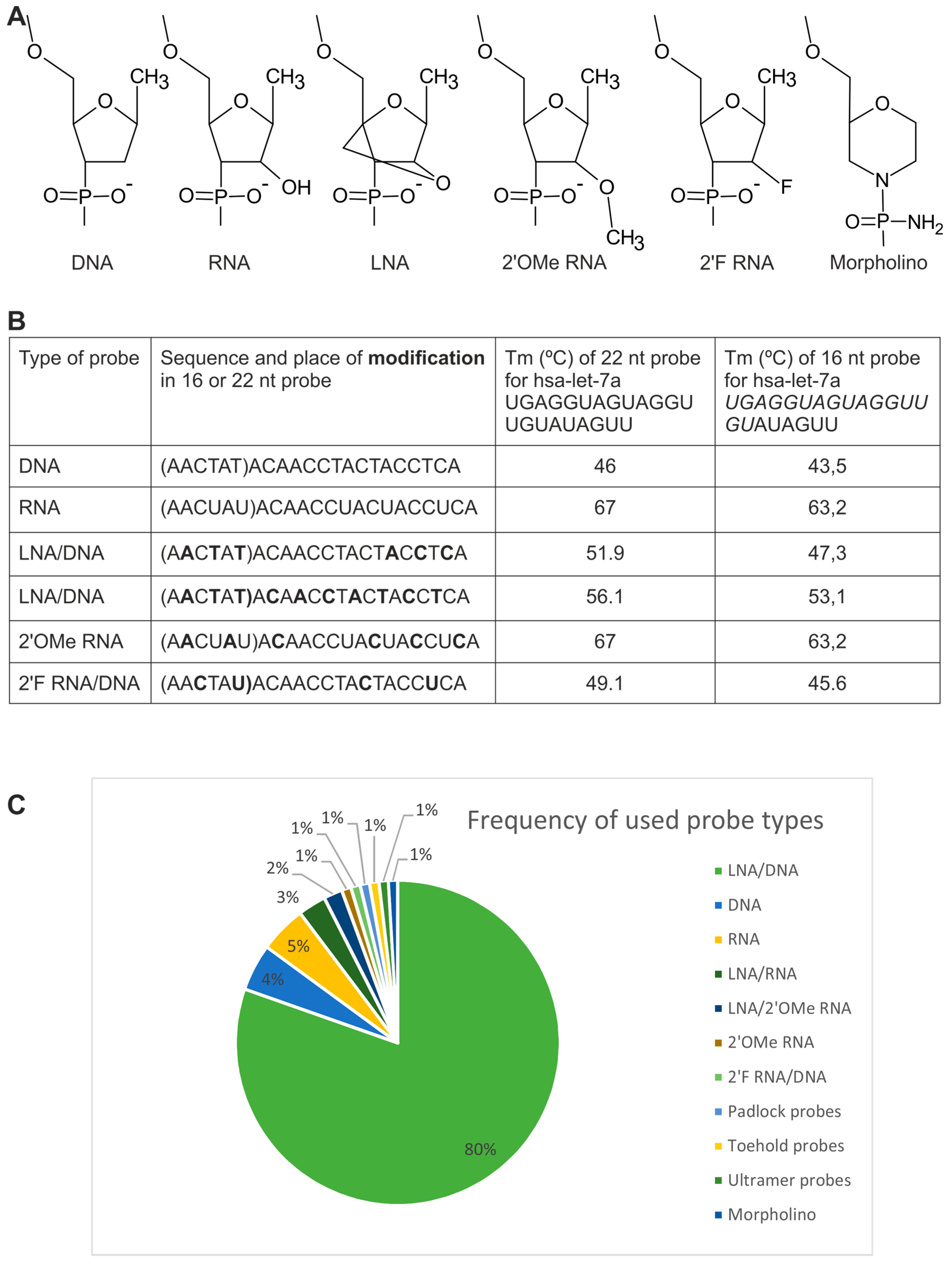
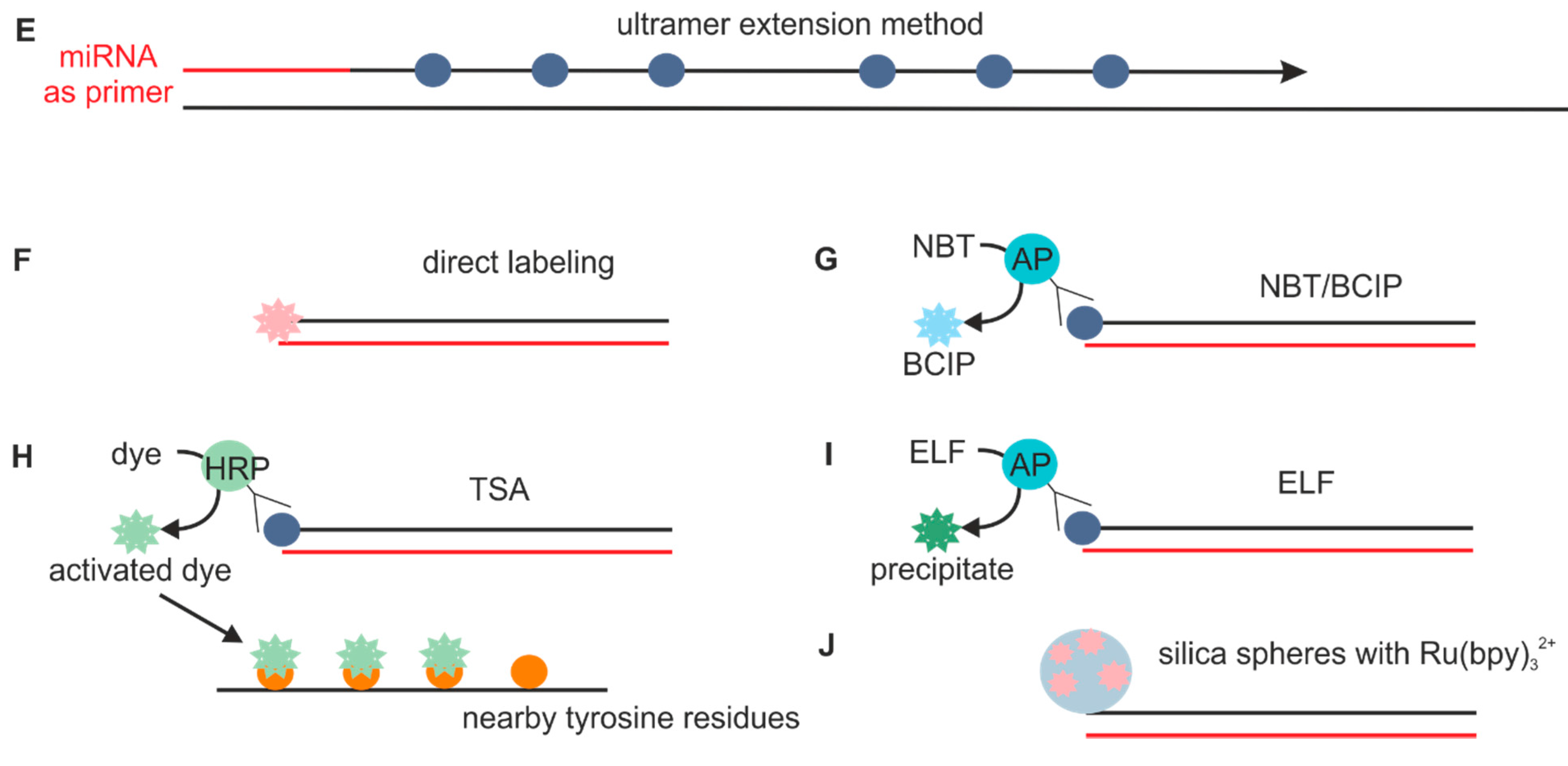
2.1.2. Probes Used with the Sequence Amplification System
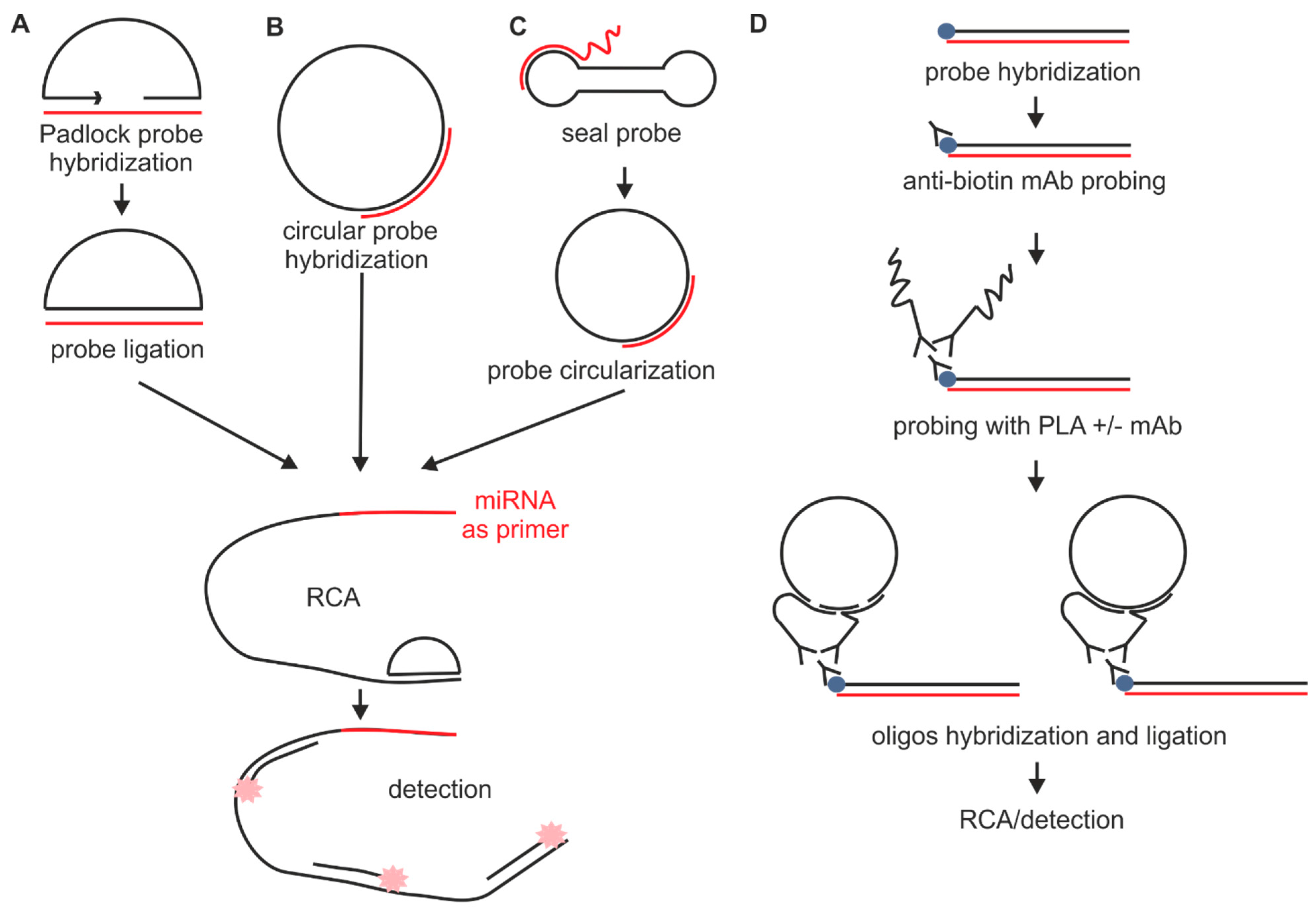
2.2. Fixation
2.3. Permeabilization
2.4. Hybridization
2.5. Post-Hybridization Washing
2.6. Sequence Amplification Methods
2.7. Detection
2.8. Specificity Controls
3. Applications of Small RNA ISH
3.1. Small RNAs in Tissues: Presence and Expression Levels
| Probe Type | Detection Method | Cell Lines | Cryosections | Paraffinic Tissue Sections | miRNA | siRNA | piRNA | Multiplex miRNA ISH | References |
|---|---|---|---|---|---|---|---|---|---|
| LNA/DNA probes | TSA | + | + | + | + | + | - | YES | [10,22,25,30,31,107,115] |
| LNA/DNA probes | AP | + | + | + | + | + | + | YES | [20,33,87,100,116] |
| LNA/DNA probes | direct or antibody-based fluorescent detection | + | − | − | + | − | + | YES | [94,96,103] |
| LNA/DNA probes | ELF | + | − | + | + | − | − | NO | [41,49] |
| DNA probes | RCA | + | − | − | + | − | − | NO | [44,45] |
3.2. Small RNAs in Cells: Subcellular Localization
3.3. Small RNAs in Complexes: Co-Localization, Correlations and Interactions with DNA, RNA and Proteins
4. Final Remarks and Future Perspectives
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Stroynowska-Czerwinska, A.; Fiszer, A.; Krzyzosiak, W.J. The panorama of miRNA-mediated mechanisms in mammalian cells. Cell. Mol. Life Sci. CMLS 2014, 71, 2253–2270. [Google Scholar] [CrossRef] [PubMed]
- Beyret, E.; Lin, H. Pinpointing the expression of piRNAs and function of the PIWI protein subfamily during spermatogenesis in the mouse. Dev. Biol. 2011, 355, 215–226. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekar, V.; Dreyer, J.L. microRNAs miR-124, let-7d and miR-181a regulate cocaine-induced plasticity. Mol. Cell. Neurosci. 2009, 42, 350–362. [Google Scholar] [CrossRef] [PubMed]
- Hollander, J.A.; Im, H.I.; Amelio, A.L.; Kocerha, J.; Bali, P.; Lu, Q.; Willoughby, D.; Wahlestedt, C.; Conkright, M.D.; Kenny, P.J. Striatal microRNA controls cocaine intake through CREB signalling. Nature 2010, 466, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Politz, J.C.; Zhang, F.; Pederson, T. MicroRNA-206 colocalizes with ribosome-rich regions in both the nucleolus and cytoplasm of rat myogenic cells. Proc. Natl. Acad. Sci. USA 2006, 103, 18957–18962. [Google Scholar] [CrossRef] [PubMed]
- Song, R.; Ro, S.; Michaels, J.D.; Park, C.; McCarrey, J.R.; Yan, W. Many X-linked microRNAs escape meiotic sex chromosome inactivation. Nat. Genet. 2009, 41, 488–493. [Google Scholar] [CrossRef] [PubMed]
- Sweetman, D.; Rathjen, T.; Jefferson, M.; Wheeler, G.; Smith, T.G.; Wheeler, G.N.; Munsterberg, A.; Dalmay, T. FGF-4 signaling is involved in mir-206 expression in developing somites of chicken embryos. Dev. Dyn. 2006, 235, 2185–2191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tuddenham, L.; Wheeler, G.; Ntounia-Fousara, S.; Waters, J.; Hajihosseini, M.K.; Clark, I.; Dalmay, T. The cartilage specific microRNA-140 targets histone deacetylase 4 in mouse cells. FEBS Lett. 2006, 580, 4214–4217. [Google Scholar] [CrossRef] [PubMed]
- Wienholds, E.; Kloosterman, W.P.; Miska, E.; Alvarez-Saavedra, E.; Berezikov, E.; de Bruijn, E.; Horvitz, H.R.; Kauppinen, S.; Plasterk, R.H. MicroRNA expression in zebrafish embryonic development. Science 2005, 309, 310–301. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, A.D.; Yelamanchili, S.V.; Marcondes, M.C.; Fox, H.S. Up-regulation of microRNA-142 in simian immunodeficiency virus encephalitis leads to repression of sirtuin1. FASEB J. 2013, 27, 3720–3729. [Google Scholar] [CrossRef] [PubMed]
- Gambardella, S.; Rinaldi, F.; Lepore, S.M.; Viola, A.; Loro, E.; Angelini, C.; Vergani, L.; Novelli, G.; Botta, A. Overexpression of microRNA-206 in the skeletal muscle from myotonic dystrophy type 1 patients. J. Transl. Med. 2010, 8. [Google Scholar] [CrossRef] [PubMed]
- Kluiver, J.; Poppema, S.; de Jong, D.; Blokzijl, T.; Harms, G.; Jacobs, S.; Kroesen, B.J.; van den Berg, A. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J. Pathol. 2005, 207, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.; Yang, H.; He, L.; Zhao, J.J.; Coppola, D.; Dalton, W.S.; Cheng, J.Q. MicroRNA-155 is regulated by the transforming growth factor β/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol. Cell. Biol. 2008, 28, 6773–6784. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Liu, C.; Zhu, J.; Shu, P.; Yin, B.; Gong, Y.; Qiang, B.; Yuan, J.; Peng, X. MicroRNA-16 targets amyloid precursor protein to potentially modulate Alzheimer’s-associated pathogenesis in SAMP8 mice. Neurobiol. Aging 2012, 33, 522–534. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Wang, Y.; Minto, A.W.; Wang, J.; Shi, Q.; Li, X.; Quigg, R.J. MicroRNA-377 is up-regulated and can lead to increased fibronectin production in diabetic nephropathy. FASEB J. 2008, 22, 4126–4135. [Google Scholar] [CrossRef] [PubMed]
- Graveel, C.R.; Calderone, H.M.; Westerhuis, J.J.; Winn, M.E.; Sempere, L.F. Critical analysis of the potential for microRNA biomarkers in breast cancer management. Breast Cancer 2015, 7, 59–79. [Google Scholar] [PubMed]
- Tian, T.; Wang, J.; Zhou, X. A review: MicroRNA detection methods. Org. Biomol. Chem. 2015, 13, 2226–2238. [Google Scholar] [CrossRef] [PubMed]
- Levsky, J.M.; Singer, R.H. Fluorescence in situ hybridization: Past, present and future. J. Cell Sci. 2003, 116, 2833–2838. [Google Scholar] [CrossRef] [PubMed]
- Donnem, T.; Eklo, K.; Berg, T.; Sorbye, S.W.; Lonvik, K.; Al-Saad, S.; Al-Shibli, K.; Andersen, S.; Stenvold, H.; Bremnes, R.M. Prognostic impact of miR-155 in non-small cell lung cancer evaluated by in situ hybridization. J. Transl. Med. 2011, 9. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, B.S.; Jorgensen, S.; Fog, J.U.; Sokilde, R.; Christensen, I.J.; Hansen, U.; Brunner, N.; Baker, A.; Moller, S.; Nielsen, H.J. High levels of microRNA-21 in the stroma of colorectal cancers predict short disease-free survival in stage II colon cancer patients. Clin. Exp. Metastasis 2011, 28, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Pena, J.T.; Sohn-Lee, C.; Rouhanifard, S.H.; Ludwig, J.; Hafner, M.; Mihailovic, A.; Lim, C.; Holoch, D.; Berninger, P.; Zavolan, M.; et al. miRNA in situ hybridization in formaldehyde and EDC-fixed tissues. Nat. Methods 2009, 6, 139–141. [Google Scholar] [CrossRef] [PubMed]
- Renwick, N.; Cekan, P.; Masry, P.A.; McGeary, S.E.; Miller, J.B.; Hafner, M.; Li, Z.; Mihailovic, A.; Morozov, P.; Brown, M.; et al. Multicolor microRNA FISH effectively differentiates tumor types. J. Clin. Investig. 2013, 123, 2694–2702. [Google Scholar] [CrossRef] [PubMed]
- Yamamichi, N.; Shimomura, R.; Inada, K.; Sakurai, K.; Haraguchi, T.; Ozaki, Y.; Fujita, S.; Mizutani, T.; Furukawa, C.; Fujishiro, M.; et al. Locked nucleic acid in situ hybridization analysis of miR-21 expression during colorectal cancer development. Clin. Cancer Res. 2009, 15, 4009–4016. [Google Scholar] [CrossRef] [PubMed]
- Politz, J.C.; Hogan, E.M.; Pederson, T. MicroRNAs with a nucleolar location. RNA 2009, 15, 1705–1715. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Johnson, J.J.; Stack, M.S. Fluorescence in situ hybridization for microRNA Detection in Archived Oral Cancer Tissues. J. Oncol. 2012, 2012. [Google Scholar] [CrossRef] [PubMed]
- Xu, N.; Papagiannakopoulos, T.; Pan, G.; Thomson, J.A.; Kosik, K.S. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell 2009, 137, 647–658. [Google Scholar] [CrossRef] [PubMed]
- Dixon-McIver, A.; East, P.; Mein, C.A.; Cazier, J.B.; Molloy, G.; Chaplin, T.; Andrew Lister, T.; Young, B.D.; Debernardi, S. Distinctive patterns of microRNA expression associated with karyotype in acute myeloid leukaemia. PLoS ONE 2008, 3, e2141. [Google Scholar] [CrossRef] [PubMed]
- Majid, S.; Dar, A.A.; Saini, S.; Yamamura, S.; Hirata, H.; Tanaka, Y.; Deng, G.; Dahiya, R. MicroRNA-205-directed transcriptional activation of tumor suppressor genes in prostate cancer. Cancer 2010, 116, 5637–5649. [Google Scholar] [CrossRef] [PubMed]
- Morton, S.U.; Scherz, P.J.; Cordes, K.R.; Ivey, K.N.; Stainier, D.Y.; Srivastava, D. microRNA-138 modulates cardiac patterning during embryonic development. Proc. Natl. Acad. Sci. USA 2008, 105, 17830–17835. [Google Scholar] [CrossRef] [PubMed]
- Quesne, J.L.; Jones, J.; Warren, J.; Dawson, S.J.; Ali, H.R.; Bardwell, H.; Blows, F.; Pharoah, P.; Caldas, C. Biological and prognostic associations of miR-205 and let-7b in breast cancer revealed by in situ hybridization analysis of micro-RNA expression in arrays of archival tumour tissue. J. Pathol. 2012, 227, 306–314. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Johnson, J.J.; Stack, M.S. Detecting microRNA in human cancer tissues with fluorescence in situ hybridization. Methods Mol. Biol. 2013, 1039, 19–27. [Google Scholar] [PubMed]
- Song, Y.X.; Yue, Z.Y.; Wang, Z.N.; Xu, Y.Y.; Luo, Y.; Xu, H.M.; Zhang, X.; Jiang, L.; Xing, C.Z.; Zhang, Y. MicroRNA-148b is frequently down-regulated in gastric cancer and acts as a tumor suppressor by inhibiting cell proliferation. Mol. Cancer 2011, 10. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.L.; Ohtsuka, T.; Gonzalez, A.; Kageyama, R. MicroRNA9 regulates neural stem cell differentiation by controlling Hes1 expression dynamics in the developing brain. Genes Cells Devoted Mol. Cell. Mech. 2012, 17, 952–961. [Google Scholar] [CrossRef] [PubMed]
- Wiklund, E.D.; Bramsen, J.B.; Hulf, T.; Dyrskjot, L.; Ramanathan, R.; Hansen, T.B.; Villadsen, S.B.; Gao, S.; Ostenfeld, M.S.; Borre, M.; et al. Coordinated epigenetic repression of the miR-200 family and miR-205 in invasive bladder cancer. Int. J. Cancer 2011, 128, 1327–1334. [Google Scholar] [CrossRef] [PubMed]
- Yeh, Y.M.; Chuang, C.M.; Chao, K.C.; Wang, L.H. MicroRNA-138 suppresses ovarian cancer cell invasion and metastasis by targeting SOX4 and HIF-1α. Int. J. Cancer 2013, 133, 867–878. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.Z.; Zhang, J.X.; Zhang, A.L.; Shi, Z.D.; Han, L.; Jia, Z.F.; Yang, W.D.; Wang, G.X.; Jiang, T.; You, Y.P.; et al. MiR-221 and miR-222 target PUMA to induce cell survival in glioblastoma. Mol. Cancer 2010, 9. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulou, A.S.; Serneels, L.; Achsel, T.; Mandemakers, W.; Callaerts-Vegh, Z.; Dooley, J.; Lau, P.; Ayoubi, T.; Radaelli, E.; Spinazzi, M. Deficiency of the miR-29a/b-1 cluster leads to ataxic features and cerebellar alterations in mice. Neurobiol. Dis. 2014, 73c, 275–288. [Google Scholar] [CrossRef] [PubMed]
- Soe, M.J.; Moller, T.; Dufva, M.; Holmstrom, K. A sensitive alternative for microRNA in situ hybridizations using probes of 2′-O-methyl RNA + LNA. J. Histochem. Cytochem. 2011, 59, 661–672. [Google Scholar] [CrossRef] [PubMed]
- Deo, M.; Yu, J.Y.; Chung, K.H.; Tippens, M.; Turner, D.L. Detection of mammalian microRNA expression by in situ hybridization with RNA oligonucleotides. Dev. Dyn. 2006, 235, 2538–2548. [Google Scholar] [CrossRef] [PubMed]
- Thompson, R.C.; Deo, M.; Turner, D.L. Analysis of microRNA expression by in situ hybridization with RNA oligonucleotide probes. Methods 2007, 43, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, X.; Li, Y.; Yang, H.; Wang, L.; Qin, Y.; Liu, H.; Fu, L.; Guan, X.Y. Cell-specific detection of miR-375 downregulation for predicting the prognosis of esophageal squamous cell carcinoma by miRNA in situ hybridization. PLoS ONE 2013, 8, e53582. [Google Scholar] [CrossRef] [PubMed]
- Lagendijk, A.K.; Moulton, J.D.; Bakkers, J. Revealing details: Whole mount microRNA in situ hybridization protocol for zebrafish embryos and adult tissues. Biol. Open 2012, 1, 566–569. [Google Scholar] [CrossRef] [PubMed]
- Jonstrup, S.P.; Koch, J.; Kjems, J. A microRNA detection system based on padlock probes and rolling circle amplification. RNA 2006, 12, 1747–1752. [Google Scholar] [CrossRef] [PubMed]
- Jones, K.L.; Karpala, A.; Hirst, B.; Jenkins, K.; Tizard, M.; Pereira, C.F.; Leis, A.; Monaghan, P.; Hyatt, A.; Mak, J. Visualising single molecules of HIV-1 and miRNA nucleic acids. BMC Cell Biol. 2013, 14. [Google Scholar] [CrossRef] [PubMed]
- Ge, J.; Zhang, L.L.; Liu, S.J.; Yu, R.Q.; Chu, X. A highly sensitive target-primed rolling circle amplification (TPRCA) method for fluorescent in situ hybridization detection of microRNA in tumor cells. Anal. Chem. 2014, 86, 1808–1815. [Google Scholar] [CrossRef] [PubMed]
- Deng, R.; Tang, L.; Tian, Q.; Wang, Y.; Lin, L.; Li, J. Toehold-initiated rolling circle amplification for visualizing individual microRNAs in situ in single cells. Angew. Chem. Int. Ed. Engl. 2014, 53, 2389–2393. [Google Scholar] [CrossRef] [PubMed]
- Nuovo, G.J.; Elton, T.S.; Nana-Sinkam, P.; Volinia, S.; Croce, C.M.; Schmittgen, T.D. A methodology for the combined in situ analyses of the precursor and mature forms of microRNAs and correlation with their putative targets. Nat. Protoc. 2009, 4, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, A.D.; Yelamanchili, S.V.; Fox, H.S. Combined fluorescent in situ hybridization for detection of microRNAs and immunofluorescent labeling for cell-type markers. Front. Cell. Neurosci. 2013, 7. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Tsourkas, A. Imaging individual microRNAs in single mammalian cells in situ. Nucleic Acids Res. 2009, 37. [Google Scholar] [CrossRef] [PubMed]
- Darnell, D.K.; Kaur, S.; Stanislaw, S.; Konieczka, J.H.; Yatskievych, T.A.; Antin, P.B. MicroRNA expression during chick embryo development. Dev. Dyn. 2006, 235, 3156–3165. [Google Scholar] [CrossRef] [PubMed]
- Obernosterer, G.; Martinez, J.; Alenius, M. Locked nucleic acid-based in situ detection of microRNAs in mouse tissue sections. Nat. Protoc. 2007, 2, 1508–1514. [Google Scholar] [CrossRef] [PubMed]
- Wayman, G.A.; Davare, M.; Ando, H.; Fortin, D.; Varlamova, O.; Cheng, H.Y.; Marks, D.; Obrietan, K.; Soderling, T.R.; Goodman, R.H.; et al. An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc. Natl. Acad. Sci. USA 2008, 105, 9093–9098. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, G.; Ntounia-Fousara, S.; Granda, B.; Rathjen, T.; Dalmay, T. Identification of new central nervous system specific mouse microRNAs. FEBS Lett. 2006, 580, 2195–2200. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Zou, Q.; Wang, S.P.; Tang, S.M.; Zhang, G.Z.; Shen, X.J. The discovery approaches and detection methods of microRNAs. Mol. Biol. Rep. 2011, 38, 4125–4135. [Google Scholar] [CrossRef] [PubMed]
- Kloosterman, W.P.; Wienholds, E.; de Bruijn, E.; Kauppinen, S.; Plasterk, R.H. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat. Methods 2006, 3, 27–29. [Google Scholar] [CrossRef] [PubMed]
- De Planell-Saguer, M.; Rodicio, M.C. Detection methods for microRNAs in clinic practice. Clin. Biochem. 2013, 46, 869–878. [Google Scholar] [CrossRef] [PubMed]
- Trevisan, S.; Nonis, A.; Begheldo, M.; Manoli, A.; Palme, K.; Caporale, G.; Ruperti, B.; Quaggiotti, S. Expression and tissue-specific localization of nitrate-responsive miRNAs in roots of maize seedlings. Plant Cell Environ. 2012, 35, 1137–1155. [Google Scholar] [CrossRef] [PubMed]
- Keiser, M.S.; Geoghegan, J.C.; Boudreau, R.L.; Lennox, K.A.; Davidson, B.L. RNAi or overexpression: Alternative therapies for Spinocerebellar Ataxia Type 1. Neurobiol. Dis. 2013, 56, 6–13. [Google Scholar] [CrossRef] [PubMed]
- Majlessi, M.; Nelson, N.C.; Becker, M.M. Advantages of 2′-O-methyl oligoribonucleotide probes for detecting RNA targets. Nucleic Acids Res. 1998, 26, 2224–2229. [Google Scholar] [CrossRef] [PubMed]
- Larsson, C.; Grundberg, I.; Soderberg, O.; Nilsson, M. In situ detection and genotyping of individual mRNA molecules. Nat. Methods 2010, 7, 395–397. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.S.; Wang, H.Y.; Liao, Y.C.; Tsai, P.C.; Chen, K.C.; Cheng, H.Y.; Lin, R.T.; Juo, S.H. MicroRNA-195 regulates vascular smooth muscle cell phenotype and prevents neointimal formation. Cardiovasc. Res. 2012, 95, 517–526. [Google Scholar] [CrossRef] [PubMed]
- Renwick, N.; Cekan, P.; Bognanni, C.; Tuschl, T. Multiplexed miRNA fluorescence in situ hybridization for formalin-fixed paraffin-embedded tissues. Methods Mol. Biol. 2014, 1211, 171–187. [Google Scholar] [PubMed]
- Jamur, M.C.; Oliver, C. Permeabilization of cell membranes. Methods Mol. Biol. 2010, 588, 63–66. [Google Scholar] [PubMed]
- Turnock-Jones, J.J.; le Quesne, J.P. MicroRNA in situ hybridization in tissue microarrays. Methods Mol. Biol. 2014, 1211, 85–93. [Google Scholar] [PubMed]
- Fuentes, R.; Fernandez, J. Fixation/permeabilization procedure for mRNA in situ hybridization of zebrafish whole-mount oocytes, embryos, and larvae. Methods Mol. Biol. 2014, 1211, 1–13. [Google Scholar] [PubMed]
- Fernandez, J.; Fuentes, R. Fixation/permeabilization: New alternative procedure for immunofluorescence and mRNA in situ hybridization of vertebrate and invertebrate embryos. Dev. Dyn. 2013, 242, 503–517. [Google Scholar] [CrossRef] [PubMed]
- Ortega, F.G.; Lorente, J.A.; Garcia Puche, J.L.; Ruiz, M.P.; Sanchez-Martin, R.M.; de Miguel-Perez, D.; Diaz-Mochon, J.J.; Serrano, M.J. miRNA in situ hybridization in circulating tumor cells—MishCTC. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Kosman, D.; Mizutani, C.M.; Lemons, D.; Cox, W.G.; McGinnis, W.; Bier, E. Multiplex detection of RNA expression in Drosophila embryos. Science 2004, 305. [Google Scholar] [CrossRef] [PubMed]
- Sempere, L.F. Fully automated fluorescence-based four-color multiplex assay for co-detection of microRNA and protein biomarkers in clinical tissue specimens. Methods Mol. Biol. 2014, 1211, 151–170. [Google Scholar] [PubMed]
- Zhang, J.; Fu, Y.; Mei, Y.; Jiang, F.; Lakowicz, J.R. Fluorescent metal nanoshell probe to detect single miRNA in lung cancer cell. Anal. Chem. 2010, 82, 4464–4471. [Google Scholar] [CrossRef] [PubMed]
- Houwing, S.; Kamminga, L.M.; Berezikov, E.; Cronembold, D.; Girard, A.; van den Elst, H.; Filippov, D.V.; Blaser, H.; Raz, E.; Moens, C.B.; et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell 2007, 129, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, S.; Baker, A.; Moller, S.; Nielsen, B.S. Robust one-day in situ hybridization protocol for detection of microRNAs in paraffin samples using LNA probes. Methods 2010, 52, 375–381. [Google Scholar] [CrossRef] [PubMed]
- Nuovo, G.J. In situ detection of microRNAs in paraffin embedded, formalin fixed tissues and the co-localization of their putative targets. Methods 2010, 52, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Speel, E.J.; Hopman, A.H.; Komminoth, P. Amplification methods to increase the sensitivity of in situ hybridization: Play card(s). J. Histochem. Cytochem. 1999, 47, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yang, B. Quantitative LNA-ELF-FISH method for miRNA detection in single mammalian cell. In MicroRNA Expression Detection Methods; Springer: Berlin, Germany; Heidelberg, Germany, 2010; pp. 353–359. [Google Scholar]
- Lin, S.L.; Chiang, A.; Chang, D.; Ying, S.Y. Loss of miR-146a function in hormone-refractory prostate cancer. RNA 2008, 14, 417–424. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Nan, Y.; Han, L.; Zhang, A.; Wang, G.; Jia, Z.; Hao, J.; Pu, P.; Zhong, Y.; Kang, C. MicroRNA miR-451 downregulates the PI3K/AKT pathway through CAB39 in human glioma. Int. J. Oncol. 2012, 40, 1105–1112. [Google Scholar] [PubMed]
- You, X.; Vlatkovic, I.; Babic, A.; Will, T.; Epstein, I.; Tushev, G.; Akbalik, G.; Wang, M.; Glock, C.; Quedenau, C.; et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat. Neurosci. 2015, 18, 603–610. [Google Scholar] [CrossRef] [PubMed]
- Silahtaroglu, A.N.; Nolting, D.; Dyrskjot, L.; Berezikov, E.; Moller, M.; Tommerup, N.; Kauppinen, S. Detection of microRNAs in frozen tissue sections by fluorescence in situ hybridization using locked nucleic acid probes and tyramide signal amplification. Nat. Protoc. 2007, 2, 2520–2528. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Khanna, S.; Hussain, S.R.; Biswas, S.; Azad, A.; Rink, C.; Gnyawali, S.; Shilo, S.; Nuovo, G.J.; Sen, C.K. MicroRNA expression in response to murine myocardial infarction: miR-21 regulates fibroblast metalloprotease-2 via phosphatase and tensin homologue. Cardiovasc. Res. 2009, 82, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Kuwabara, Y.; Ono, K.; Horie, T.; Nishi, H.; Nagao, K.; Kinoshita, M.; Watanabe, S.; Baba, O.; Kojima, Y.; Shizuta, S.; et al. Increased microRNA-1 and microRNA-133a levels in serum of patients with cardiovascular disease indicate myocardial damage. Circ. Cardiovasc. Genet. 2011, 4, 446–454. [Google Scholar] [CrossRef] [PubMed]
- Torella, D.; Iaconetti, C.; Catalucci, D.; Ellison, G.M.; Leone, A.; Waring, C.D.; Bochicchio, A.; Vicinanza, C.; Aquila, I.; Curcio, A.; et al. MicroRNA-133 controls vascular smooth muscle cell phenotypic switch in vitro and vascular remodeling in vivo. Circ. Res. 2011, 109, 880–893. [Google Scholar] [CrossRef] [PubMed]
- Kocerha, J.; Kauppinen, S.; Wahlestedt, C. microRNAs in CNS disorders. Neuromol. Med. 2009, 11, 162–172. [Google Scholar] [CrossRef] [PubMed]
- Hebert, S.S.; Horre, K.; Nicolai, L.; Papadopoulou, A.S.; Mandemakers, W.; Silahtaroglu, A.N.; Kauppinen, S.; Delacourte, A.; de Strooper, B. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer’s disease correlates with increased BACE1/β-secretase expression. Proc. Natl. Acad. Sci. USA 2008, 105, 6415–6420. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.X.; Wilfred, B.R.; Madathil, S.K.; Tang, G.; Hu, Y.; Dimayuga, J.; Stromberg, A.J.; Huang, Q.; Saatman, K.E.; Nelson, P.T. miR-107 regulates granulin/progranulin with implications for traumatic brain injury and neurodegenerative disease. Am. J. Pathol. 2010, 177, 334–345. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Guan, Y.; Chen, Y.; Zhang, C.; Yu, L.; Gao, H.; Du, H.; Liu, B.; Wang, X. miRNA-9 expression is upregulated in the spinal cord of G93A-SOD1 transgenic mice. Int. J. Clin. Exp. Pathol. 2013, 6, 1826–1838. [Google Scholar] [PubMed]
- De Felice, B.; Annunziata, A.; Fiorentino, G.; Borra, M.; Biffali, E.; Coppola, C.; Cotrufo, R.; Brettschneider, J.; Giordana, M.L.; Dalmay, T.; et al. miR-338-3p is over-expressed in blood, CFS, serum and spinal cord from sporadic amyotrophic lateral sclerosis patients. Neurogenetics 2014, 15, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Koscianska, E.; Krzyzosiak, W.J. Current understanding of the role of microRNAs in spinocerebellar ataxias. Cerebellum Ataxias 2014, 1, 7. [Google Scholar] [CrossRef]
- Koscianska, E.K.E.; Jaworska, E.; Krzyzosiak, W.J. MicroRNA deregulation in trinucleotide repeat expansion disorders. In Applied RNAi: From Fundamental Research to Therapeutic Applications; Patrick, A., Weinberg, M.S., Eds.; Caister Academic Press: Poole, UK, 2014; pp. 227–246. [Google Scholar]
- Wang, Y.X.; Lang, F.; Liu, Y.X.; Yang, C.Q.; Gao, H.J. In situ hybridization analysis of the expression of miR-106b in colonic cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 786–792. [Google Scholar] [PubMed]
- Kjaer-Frifeldt, S.; Hansen, T.F.; Nielsen, B.S.; Joergensen, S.; Lindebjerg, J.; Soerensen, F.B.; dePont Christensen, R.; Jakobsen, A. The prognostic importance of miR-21 in stage II colon cancer: A population-based study. Br. J. Cancer 2012, 107, 1169–1174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nielsen, B.S.; Holmstrom, K. Combined microRNA in situ hybridization and immunohistochemical detection of protein markers. Methods Mol. Biol. 2013, 986, 353–365. [Google Scholar] [PubMed]
- Kapsimali, M.; Kloosterman, W.P.; de Bruijn, E.; Rosa, F.; Plasterk, R.H.; Wilson, S.W. MicroRNAs show a wide diversity of expression profiles in the developing and mature central nervous system. Genome Biol. 2007, 8. [Google Scholar] [CrossRef] [PubMed]
- Hansen, K.F.; Karelina, K.; Sakamoto, K.; Wayman, G.A.; Impey, S.; Obrietan, K. miRNA-132: A dynamic regulator of cognitive capacity. Brain Struct. Funct. 2013, 218, 817–831. [Google Scholar] [CrossRef] [PubMed]
- Schratt, G. microRNAs at the synapse. Nat. Rev. Neurosci. 2009, 10, 842–849. [Google Scholar] [CrossRef] [PubMed]
- Cong, N.; Du, P.; Zhang, A.; Shen, F.; Su, J.; Pu, P.; Wang, T.; Zjang, J.; Kang, C.; Zhang, Q. Downregulated microRNA-200a promotes EMT and tumor growth through the Wnt/β-catenin pathway by targeting the E-cadherin repressors ZEB1/ZEB2 in gastric adenocarcinoma. Oncol. Rep. 2013, 29, 1579–1587. [Google Scholar] [PubMed]
- Kong, W.; He, L.; Coppola, M.; Guo, J.; Esposito, N.N.; Coppola, D.; Cheng, J.Q. MicroRNA-155 regulates cell survival, growth, and chemosensitivity by targeting FOXO3a in breast cancer. J. Biol. Chem. 2010, 285, 17869–17879. [Google Scholar] [CrossRef] [PubMed]
- Lodygin, D.; Tarasov, V.; Epanchintsev, A.; Berking, C.; Knyazeva, T.; Korner, H.; Knyazev, P.; Diebold, J.; Hermeking, H. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle 2008, 7, 2591–2600. [Google Scholar] [CrossRef] [PubMed]
- Dillhoff, M.; Liu, J.; Frankel, W.; Croce, C.; Bloomston, M. MicroRNA-21 is overexpressed in pancreatic cancer and a potential predictor of survival. J. Gastrointest. Surg. 2008, 12, 2171–2176. [Google Scholar] [CrossRef] [PubMed]
- Hermansen, S.K.; Dahlrot, R.H.; Nielsen, B.S.; Hansen, S.; Kristensen, B.W. miR-21 expression in the tumor cell compartment holds unfavorable prognostic value in gliomas. J. Neuro-Oncol. 2013, 111, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Oom, A.L.; Humphries, B.A.; Yang, C. MicroRNAs: Novel players in cancer diagnosis and therapies. 2014; 2014. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, K.Y.; Liu, S.M.; Sen, S. Tumor-associated circulating microRNAs as biomarkers of cancer. Molecules 2014, 19, 1912–1938. [Google Scholar] [CrossRef] [PubMed]
- Ning, X.; Shi, Z.; Liu, X.; Zhang, A.; Han, L.; Jiang, K.; Kang, C.; Zhang, Q. DNMT1 and EZH2 mediated methylation silences the microRNA-200b/a/429 gene and promotes tumor progression. Cancer Lett. 2015, 359, 198–205. [Google Scholar] [CrossRef] [PubMed]
- Schetter, A.J.; Leung, S.Y.; Sohn, J.J.; Zanetti, K.A.; Bowman, E.D.; Yanaihara, N.; Yuen, S.T.; Chan, T.L.; Kwong, D.L.; Au, G.K.; et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA 2008, 299, 425–436. [Google Scholar] [CrossRef] [PubMed]
- Rask, L.; Balslev, E.; Jorgensen, S.; Eriksen, J.; Flyger, H.; Moller, S.; Hogdall, E.; Litman, T.; Nielsen, B.S. High expression of miR-21 in tumor stroma correlates with increased cancer cell proliferation in human breast cancer. APMIS Acta Pathol. Microbiol. Immunol. Scand. 2011, 119, 663–673. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, B.S.; Balslev, E.; Poulsen, T.S.; Nielsen, D.; Moller, T.; Mortensen, C.E.; Holmstrom, K.; Hogdall, E. miR-21 expression in cancer cells may not predict resistance to adjuvant trastuzumab in primary breast cancer. Front. Oncol. 2014, 4. [Google Scholar] [CrossRef] [PubMed]
- MacKenzie, T.A.; Schwartz, G.N.; Calderone, H.M.; Graveel, C.R.; Winn, M.E.; Hostetter, G.; Wells, W.A.; Sempere, L.F. Stromal expression of miR-21 identifies high-risk group in triple-negative breast cancer. Am. J. Pathol. 2014, 184, 3217–3225. [Google Scholar] [CrossRef] [PubMed]
- Preis, M.; Gardner, T.B.; Gordon, S.R.; Pipas, J.M.; Mackenzie, T.A.; Klein, E.E.; Longnecker, D.S.; Gutmann, E.J.; Sempere, L.F.; Korc, M. MicroRNA-10b expression correlates with response to neoadjuvant therapy and survival in pancreatic ductal adenocarcinoma. Clin. Cancer Res. 2011, 17, 5812–5821. [Google Scholar] [CrossRef] [PubMed]
- Hanna, J.A.; Wimberly, H.; Kumar, S.; Slack, F.; Agarwal, S.; Rimm, D.L. Quantitative analysis of microRNAs in tissue microarrays by in situ hybridization. BioTechniques 2012, 52, 235–245. [Google Scholar] [CrossRef] [PubMed]
- Nogueira, F.T.; Madi, S.; Chitwood, D.H.; Juarez, M.T.; Timmermans, M.C. Two small regulatory RNAs establish opposing fates of a developmental axis. Genes Dev. 2007, 21, 750–755. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Hu, H.Y.; Jiang, X.; Maierhofer, V.; Neb, E.; He, L.; Hu, Y.; Hu, H.; Li, N.; Chen, W.; Khaitovich, P. Widespread expression of piRNA-like molecules in somatic tissues. Nucleic Acids Res. 2011, 39, 6596–6607. [Google Scholar] [CrossRef] [PubMed]
- Pontes, O.; Li, C.F.; Costa Nunes, P.; Haag, J.; Ream, T.; Vitins, A.; Jacobsen, S.E.; Pikaard, C.S. The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell 2006, 126, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Bak, M.; Silahtaroglu, A.; Moller, M.; Christensen, M.; Rath, M.F.; Skryabin, B.; Tommerup, N.; Kauppinen, S. MicroRNA expression in the adult mouse central nervous system. RNA (New York, NY) 2008, 14, 432–444. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.T.; Yeh, Y.M.; Chuang, C.M.; Yang, S.Y.; Chang, J.W.; Sun, S.P.; Wang, Y.S.; Chao, K.C.; Wang, L.H. Glucocorticoids mediate induction of microRNA-708 to suppress ovarian cancer metastasis through targeting Rap1B. Nat. Commun. 2015, 6, 5917. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Ferlito, M.; Kent, O.A.; Fox-Talbot, K.; Wang, R.; Liu, D.; Raghavachari, N.; Yang, Y.; Wheelan, S.J.; Murphy, E.; et al. Nuclear miRNA regulates the mitochondrial genome in the heart. Circ. Res. 2012, 110, 1596–1603. [Google Scholar] [CrossRef] [PubMed]
- Boon, R.A.; Iekushi, K.; Lechner, S.; Seeger, T.; Fischer, A.; Heydt, S.; Kaluza, D.; Treguer, K.; Carmona, G.; Bonauer, A.; et al. MicroRNA-34a regulates cardiac ageing and function. Nature 2013, 495, 107–110. [Google Scholar] [CrossRef] [PubMed]
- Kotaja, N.; Bhattacharyya, S.N.; Jaskiewicz, L.; Kimmins, S.; Parvinen, M.; Filipowicz, W.; Sassone-Corsi, P. The chromatoid body of male germ cells: Similarity with processing bodies and presence of Dicer and microRNA pathway components. Proc. Natl. Acad. Sci. USA 2006, 103, 2647–2652. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, S.N.; Habermacher, R.; Martine, U.; Closs, E.I.; Filipowicz, W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 2006, 125, 1111–1124. [Google Scholar] [CrossRef] [PubMed]
- Barrey, E.; Saint-Auret, G.; Bonnamy, B.; Damas, D.; Boyer, O.; Gidrol, X. Pre-microRNA and mature microRNA in human mitochondria. PLoS ONE 2011, 6, e20220. [Google Scholar] [CrossRef] [PubMed]
- Khudayberdiev, S.A.; Zampa, F.; Rajman, M.; Schratt, G. A comprehensive characterization of the nuclear microRNA repertoire of post-mitotic neurons. Front. Mol. Neurosci. 2013, 6. [Google Scholar] [CrossRef] [PubMed]
- Broderick, K.E.; Chan, A.; Lin, F.; Shen, X.; Kichaev, G.; Khan, A.S.; Aubin, J.; Zimmermann, T.S.; Sardesai, N.Y. Optimized in vivo transfer of small interfering RNA targeting dermal tissue using in vivo surface electroporation. Mol. Ther. Nucleic Acids 2012, 1. [Google Scholar] [CrossRef] [PubMed]
- Shi, B.; Keough, E.; Matter, A.; Leander, K.; Young, S.; Carlini, E.; Sachs, A.B.; Tao, W.; Abrams, M.; Howell, B.; et al. Biodistribution of small interfering RNA at the organ and cellular levels after lipid nanoparticle-mediated delivery. J. Histochem. Cytochem. 2011, 59, 727–740. [Google Scholar] [CrossRef] [PubMed]
- Molitoris, B.A.; Dagher, P.C.; Sandoval, R.M.; Campos, S.B.; Ashush, H.; Fridman, E.; Brafman, A.; Faerman, A.; Atkinson, S.J.; Thompson, J.D.; et al. siRNA targeted to p53 attenuates ischemic and cisplatin-induced acute kidney injury. J. Am. Soc. Nephrol. JASN 2009, 20, 1754–1764. [Google Scholar] [CrossRef] [PubMed]
- Takanashi, M.; Oikawa, K.; Sudo, K.; Tanaka, M.; Fujita, K.; Ishikawa, A.; Nakae, S.; Kaspar, R.L.; Matsuzaki, M.; Kudo, M.; et al. Therapeutic silencing of an endogenous gene by siRNA cream in an arthritis model mouse. Gene Ther. 2009, 16, 982–989. [Google Scholar] [CrossRef] [PubMed]
- Paul, C.P.; Good, P.D.; Winer, I.; Engelke, D.R. Effective expression of small interfering RNA in human cells. Nat. Biotechnol. 2002, 20, 505–508. [Google Scholar] [CrossRef] [PubMed]
- Chandramouli, A.; Onyeagucha, B.C.; Mercado-Pimentel, M.E.; Stankova, L.; Shahin, N.A.; LaFleur, B.J.; Heimark, R.L.; Bhattacharyya, A.K.; Nelson, M.A. MicroRNA-101 (miR-101) post-transcriptionally regulates the expression of EP4 receptor in colon cancers. Cancer Biol. Ther. 2012, 13, 175–183. [Google Scholar] [CrossRef] [PubMed]
- Cougot, N.; Bhattacharyya, S.N.; Tapia-Arancibia, L.; Bordonne, R.; Filipowicz, W.; Bertrand, E.; Rage, F. Dendrites of mammalian neurons contain specialized P-body-like structures that respond to neuronal activation. J. Neurosci. 2008, 28, 13793–13804. [Google Scholar] [CrossRef] [PubMed]
- Urbanek, M.O.; Galka-Marciniak, P.; Olejniczak, M.; Krzyzosiak, W.J. RNA imaging in living cells—Methods and applications. RNA Biol. 2014, 11, 1083–1095. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Spector, D.L. Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr. Biol. CB 2007, 17, 818–823. [Google Scholar] [CrossRef] [PubMed]
- Fujioka, Y.; Utsumi, M.; Ohba, Y.; Watanabe, Y. Location of a possible miRNA processing site in SmD3/SmB nuclear bodies in Arabidopsis. Plant Cell Physiol. 2007, 48, 1243–1253. [Google Scholar] [CrossRef] [PubMed]
- Liao, X.; Wang, Q.; Ju, H. Simultaneous sensing of intracellular microRNAs with a multi-functionalized carbon nitride nanosheet probe. Chem. Commun. 2014, 50, 13604–13607. [Google Scholar] [CrossRef] [PubMed]
- Femino, A.M.; Fay, F.S.; Fogarty, K.; Singer, R.H. Visualization of single RNA transcripts in situ. Science 1998, 280, 585–590. [Google Scholar] [CrossRef] [PubMed]
- Chou, Y.Y.; Heaton, N.S.; Gao, Q.; Palese, P.; Singer, R.H.; Lionnet, T. Colocalization of different influenza viral RNA segments in the cytoplasm before viral budding as shown by single-molecule sensitivity FISH analysis. PLoS Pathog. 2013, 9, e1003358. [Google Scholar] [CrossRef] [PubMed]
- Buxbaum, A.R.; Wu, B.; Singer, R.H. Single β-actin mRNA detection in neurons reveals a mechanism for regulating its translatability. Science 2014, 343, 419–422. [Google Scholar] [CrossRef] [PubMed]
- Christoffersen, N.R.; Silahtaroglu, A.; Orom, U.A.; Kauppinen, S.; Lund, A.H. miR-200b mediates post-transcriptional repression of ZFHX1B. RNA 2007, 13, 1172–1178. [Google Scholar] [CrossRef] [PubMed]
- Kucherenko, M.M.; Barth, J.; Fiala, A.; Shcherbata, H.R. Steroid-induced microRNA let-7 acts as a spatio-temporal code for neuronal cell fate in the developing Drosophila brain. EMBO J. 2012, 31, 4511–4523. [Google Scholar] [CrossRef] [PubMed]
- Kucherenko, M.M.; Marrone, A.K.; Rishko, V.M.; Yatsenko, A.S.; Klepzig, A.; Shcherbata, H.R. Paraffin-embedded and frozen sections of Drosophila adult muscles. J. Vis. Exp. 2010, 46. [Google Scholar] [CrossRef]
- Zimmerman, S.G.; Peters, N.C.; Altaras, A.E.; Berg, C.A. Optimized RNA ISH, RNA FISH and protein-RNA double labeling (IF/FISH) in Drosophila ovaries. Nat. Protoc. 2013, 8, 2158–2179. [Google Scholar] [CrossRef] [PubMed]
- Yatsenko, A.S.; Shcherbata, H.R. Drosophila miR-9a targets the ECM receptor Dystroglycan to canalize myotendinous junction formation. Dev. Cell 2014, 28, 335–348. [Google Scholar] [CrossRef] [PubMed]
- Birren, S.J.; Lo, L.; Anderson, D.J. Sympathetic neuroblasts undergo a developmental switch in trophic dependence. Development 1993, 119, 597–610. [Google Scholar] [PubMed]
- Ason, B.; Darnell, D.K.; Wittbrodt, B.; Berezikov, E.; Kloosterman, W.P.; Wittbrodt, J.; Antin, P.B.; Plasterk, R.H. Differences in vertebrate microRNA expression. Proc. Natl. Acad. Sci. USA 2006, 103, 14385–14389. [Google Scholar] [CrossRef] [PubMed]
- Weston, M.D.; Pierce, M.L.; Rocha-Sanchez, S.; Beisel, K.W.; Soukup, G.A. MicroRNA gene expression in the mouse inner ear. Brain Res. 2006, 1111, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Stark, A.; Brennecke, J.; Bushati, N.; Russell, R.B.; Cohen, S.M. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell 2005, 123, 1133–1146. [Google Scholar] [CrossRef] [PubMed]
- Nelson, P.T.; Wang, W.X.; Rajeev, B.W. MicroRNAs (miRNAs) in neurodegenerative diseases. Brain Pathol. 2008, 18, 130–138. [Google Scholar] [CrossRef] [PubMed]
- Spanagel, R.; Pendyala, G.; Abarca, C.; Zghoul, T.; Sanchis-Segura, C.; Magnone, M.C.; Lascorz, J.; Depner, M.; Holzberg, D.; Soyka, M.; et al. The clock gene Per2 influences the glutamatergic system and modulates alcohol consumption. Nat. Med. 2005, 11, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Karali, M.; Peluso, I.; Marigo, V.; Banfi, S. Identification and characterization of microRNAs expressed in the mouse eye. Investig. Ophthalmol. Vis. Sci. 2007, 48, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Bonauer, A.; Carmona, G.; Iwasaki, M.; Mione, M.; Koyanagi, M.; Fischer, A.; Burchfield, J.; Fox, H.; Doebele, C.; Ohtani, K.; et al. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science 2009, 324, 1710–1713. [Google Scholar] [CrossRef] [PubMed]
- Marcon, E.; Babak, T.; Chua, G.; Hughes, T.; Moens, P.B. miRNA and piRNA localization in the male mammalian meiotic nucleus. Chromosome Res. 2008, 16, 243–260. [Google Scholar] [CrossRef] [PubMed]
- Shimojo, H.; Ohtsuka, T.; Kageyama, R. Oscillations in notch signaling regulate maintenance of neural progenitors. Neuron 2008, 58, 52–64. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.L.; Nishi, M.; Ohtsuka, T.; Matsui, T.; Takemoto, K.; Kamio-Miura, A.; Aburatani, H.; Shinkai, Y.; Kageyama, R. Essential roles of the histone methyltransferase ESET in the epigenetic control of neural progenitor cells during development. Development 2012, 139, 3806–3816. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, M.J.; Scarlett, J.M.; Steiner, R.A. Cloning and distribution of galanin-like peptide mRNA in the hypothalamus and pituitary of the macaque. Endocrinology 2002, 143, 755–763. [Google Scholar] [CrossRef] [PubMed]
- Le, M.T.; Teh, C.; Shyh-Chang, N.; Xie, H.; Zhou, B.; Korzh, V.; Lodish, H.F.; Lim, B. MicroRNA-125b is a novel negative regulator of p53. Genes Dev. 2009, 23, 862–876. [Google Scholar] [CrossRef] [PubMed]
- Nelson, P.T.; Baldwin, D.A.; Kloosterman, W.P.; Kauppinen, S.; Plasterk, R.H.; Mourelatos, Z. RAKE and LNA-ISH reveal microRNA expression and localization in archival human brain. RNA 2006, 12, 187–191. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.X.; Rajeev, B.W.; Stromberg, A.J.; Ren, N.; Tang, G.; Huang, Q.; Rigoutsos, I.; Nelson, P.T. The expression of microRNA miR-107 decreases early in Alzheimer’s disease and may accelerate disease progression through regulation of β-site amyloid precursor protein-cleaving enzyme 1. J. Neurosci. 2008, 28, 1213–1223. [Google Scholar] [CrossRef] [PubMed]
- Nelson, P.T.; Wilfred, B.R. In situ hybridization is a necessary experimental complement to microRNA (miRNA) expression profiling in the human brain. Neurosci. Lett. 2009, 466, 69–72. [Google Scholar] [CrossRef] [PubMed]
- Hand, N.J.; Master, Z.R.; Eauclaire, S.F.; Weinblatt, D.E.; Matthews, R.P.; Friedman, J.R. The microRNA-30 family is required for vertebrate hepatobiliary development. Gastroenterology 2009, 136, 1081–1090. [Google Scholar] [CrossRef] [PubMed]
- Wibrand, K.; Messaoudi, E.; Havik, B.; Steenslid, V.; Lovlie, R.; Steen, V.M.; Bramham, C.R. Identification of genes co-upregulated with Arc during BDNF-induced long-term potentiation in adult rat dentate gyrus in vivo. Eur. J. Neurosci. 2006, 23, 1501–1511. [Google Scholar] [CrossRef] [PubMed]
- Wibrand, K.; Panja, D.; Tiron, A.; Ofte, M.L.; Skaftnesmo, K.O.; Lee, C.S.; Pena, J.T.; Tuschl, T.; Bramham, C.R. Differential regulation of mature and precursor microRNA expression by NMDA and metabotropic glutamate receptor activation during LTP in the adult dentate gyrus in vivo. Eur. J. Neurosci. 2010, 31, 636–645. [Google Scholar] [CrossRef] [PubMed]
- Schepeler, T.; Reinert, J.T.; Ostenfeld, M.S.; Christensen, L.L.; Silahtaroglu, A.N.; Dyrskjot, L.; Wiuf, C.; Sorensen, F.J.; Kruhoffer, M.; Laurberg, S.; et al. Diagnostic and prognostic microRNAs in stage II colon cancer. Cancer Res. 2008, 68, 6416–6424. [Google Scholar] [CrossRef] [PubMed]
- Nuovo, G.J. In situ detection of precursor and mature microRNAs in paraffin embedded, formalin fixed tissues and cell preparations. Methods 2008, 44, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Sempere, L.F.; Preis, M.; Yezefski, T.; Ouyang, H.; Suriawinata, A.A.; Silahtaroglu, A.; Conejo-Garcia, J.R.; Kauppinen, S.; Wells, W.; Korc, M. Fluorescence-based codetection with protein markers reveals distinct cellular compartments for altered MicroRNA expression in solid tumors. Clin. Cancer Res. 2010, 16, 4246–4255. [Google Scholar] [CrossRef] [PubMed]
- Van den Berg, A.; Kroesen, B.J.; Kooistra, K.; de Jong, D.; Briggs, J.; Blokzijl, T.; Jacobs, S.; Kluiver, J.; Diepstra, A.; Maggio, E.; et al. High expression of B-cell receptor inducible gene BIC in all subtypes of Hodgkin lymphoma. Genes Chromosome Cancer 2003, 37, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Hanna, J.A.; Hahn, L.; Agarwal, S.; Rimm, D.L. In situ measurement of miR-205 in malignant melanoma tissue supports its role as a tumor suppressor microRNA. Lab. Investig. J. Tech. Methods Pathol. 2012, 92, 1390–1397. [Google Scholar] [CrossRef] [PubMed]
- Han, C.; Liu, Y.; Wan, G.; Choi, H.J.; Zhao, L.; Ivan, C.; He, X.; Sood, A.K.; Zhang, X.; Lu, X. The RNA-binding protein DDX1 promotes primary microRNA maturation and inhibits ovarian tumor progression. Cell Rep. 2014, 8, 1447–1460. [Google Scholar] [CrossRef] [PubMed]
- Carbone, A.; Gualeni, A.; Volpi, C.; Gloghini, A. MicroRNA detection in tumor tissue by in situ hybridization. Issues 2015, 1, 28. [Google Scholar]
- Crawford, M.; Brawner, E.; Batte, K.; Yu, L.; Hunter, M.G.; Otterson, G.A.; Nuovo, G.; Marsh, C.B.; Nana-Sinkam, S.P. MicroRNA-126 inhibits invasion in non-small cell lung carcinoma cell lines. Biochem. Biophys. Res. Commun. 2008, 373, 607–612. [Google Scholar] [CrossRef] [PubMed]
- Danilova, O.V.; Paiva, C.; Kaur, P.; Kamal, A.; Sempere, L.F.; Danilov, A.V. MIR21 is differentially expressed in the lymphoid tissue and modulated by stromal signalling in chronic lymphocytic leukaemia. Br. J. Haematol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Andrew, A.S.; Marsit, C.J.; Schned, A.R.; Seigne, J.D.; Kelsey, K.T.; Moore, J.H.; Perreard, L.; Karagas, M.R.; Sempere, L.F. Expression of tumor suppressive microRNA-34a is associated with a reduced risk of bladder cancer recurrence. Int. J. Cancer 2014. [Google Scholar] [CrossRef] [PubMed]
- Wilfred, B.R.; Wang, W.X.; Nelson, P.T. Energizing miRNA research: A review of the role of miRNAs in lipid metabolism, with a prediction that miR-103/107 regulates human metabolic pathways. Mol. Genet. Metab. 2007, 91, 209–217. [Google Scholar] [CrossRef] [PubMed]
- Nelson, P.T.; Dimayuga, J.; Wilfred, B.R. MicroRNA in situ hybridization in the human entorhinal and transentorhinal cortex. Front. Hum. Neurosci. 2010, 4. [Google Scholar] [CrossRef] [PubMed]
- Kidner, C.; Timmermans, M. In situ hybridization as a tool to study the role of microRNAs in plant development. Methods Mol. Biol. (Clifton, NJ) 2006, 342, 159–179. [Google Scholar]
- Nogueira, F.T.; Chitwood, D.H.; Madi, S.; Ohtsu, K.; Schnable, P.S.; Scanlon, M.J.; Timmermans, M.C. Regulation of small RNA accumulation in the maize shoot apex. PLoS Genet. 2009, 5. [Google Scholar] [CrossRef] [PubMed]
- Chitwood, D.H.; Nogueira, F.T.; Howell, M.D.; Montgomery, T.A.; Carrington, J.C.; Timmermans, M.C. Pattern formation via small RNA mobility. Genes Dev. 2009, 23, 549–554. [Google Scholar] [CrossRef] [PubMed]
- Douglas, R.N.; Wiley, D.; Sarkar, A.; Springer, N.; Timmermans, M.C.; Scanlon, M.J. Ragged seedling2 Encodes an ARGONAUTE7-like protein required for mediolateral expansion, but not dorsiventrality, of maize leaves. Plant Cell 2010, 22, 1441–1451. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.J.; Banerjee, S.; Zhou, H.; Jammalamadaka, A.; Arcila, M.; Manjunath, B.S.; Kosik, K.S. Identification of piRNAs in the central nervous system. RNA 2011, 17, 1090–1099. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Li, C.; Zhang, K.; Sun, H.; Tao, D.; Liu, Y.; Zhang, S.; Ma, Y. Identification of piRNAs in Hela cells by massive parallel sequencing. BMB Rep. 2010, 43, 635–641. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Urbanek, M.O.; Nawrocka, A.U.; Krzyzosiak, W.J. Small RNA Detection by in Situ Hybridization Methods. Int. J. Mol. Sci. 2015, 16, 13259-13286. https://doi.org/10.3390/ijms160613259
Urbanek MO, Nawrocka AU, Krzyzosiak WJ. Small RNA Detection by in Situ Hybridization Methods. International Journal of Molecular Sciences. 2015; 16(6):13259-13286. https://doi.org/10.3390/ijms160613259
Chicago/Turabian StyleUrbanek, Martyna O., Anna U. Nawrocka, and Wlodzimierz J. Krzyzosiak. 2015. "Small RNA Detection by in Situ Hybridization Methods" International Journal of Molecular Sciences 16, no. 6: 13259-13286. https://doi.org/10.3390/ijms160613259





