Synthesis, Characterization and Biological Evaluation of Transition Metal Complexes Derived from N, S Bidentate Ligands
Abstract
:1. Introduction
2. Results and Discussion
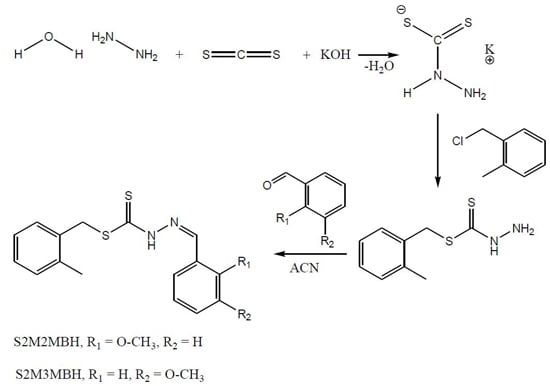
2.1. Synthesis

| Serial Number | Compound | % Found (Calculated) | |||
|---|---|---|---|---|---|
| C | H | N | M | ||
| 1 | S2M2MBH | 62.51 (61.79) | 5.44 (5.49) | 8.00 (8.48) | - |
| 2 | S2M3MBH | 64.06 (61.79) | 5.50 (5.49) | 9.08 (8.48) | - |
| 3 | [Cu(S2M2MB)2] | 57.18 (56.52) | 4.80 (4.74) | 8.20 (7.75) | 8.40 (8.8) |
| 4 | [Ni(S2M2MB)2] | 56.55 (56.91) | 4.82 (4.78) | 8.21 (7.81) | 8.54 (8.18) |
| 5 | [Zn(S2M2MB)2] | 57.34 (56.38) | 4.80 (4.73) | 8.24 (7.74) | 9.02 (9.03) |
| 6 | [Cu(S2M3MB)2] | 56.32 (56.52) | 4.58 (4.74) | 7.94 (7.75) | 8.30 (8.8) |
| 7 | [Ni(S2M3MB)2] | 56.96 (56.91) | 4.76 (4.78) | 8.21 (7.81) | 8.51 (8.18) |
| 8 | [Zn(S2M3MB)2] | 55.46 (56.38) | 4.65 (4.73) | 7.88 (7.74) | 8.79 (9.03) |
2.2. 1H- and 13C-NMR Spectra
| Compound | 13C-NMR Assignment, δ (ppm) | 1H-NMR Assignment, δ (ppm) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| –C=S | –CH3 | –CH2 | –CH | O–CH3 | Aromatic Carbons | NH | CH | CH2 | CH3 | Aromatic Protons | |
| 1 | 196.6 | 19.4 | 38.0 | 158.9 | 55.7 | 112.5–142.8 | s, 1H (13.3) | s, 1H (7.7) | s, 2H (4.4) | s, 3H (3.8 & 2.3) | d, 1H (7.2, 7.3, 7.7, 7.1) t, 1H (7.1, 7.1, 7.4, 7.0) |
| 2 | 196.1 | 19.4 | 36.8 | 162.0 | 55.9 | 115.1–147.1 | s, 1H (13.2) | s, 1H (8.3) | s, 2H (4.4) | s, 3H (3.7 & 2.3) | s, 1H (8.1) d, 1H (7.0, 7.2, 7.3, 7.6) t, 1H (7.1, 7.1, 7.1) |
2.3. IR Spectra
| Compound | IR Bands (cm−1) | µeff (BM) | Electronic Spectra (in DMSO) λmax(log εmax) | |||
|---|---|---|---|---|---|---|
| v(NH) | v(C=N) | v(N–N) | v(C=S)/v(C–S) | |||
| 1 | 3085 (w) | 1598 (m) | 1035 (s) | 736 (s) | - | 352(4.86) |
| 2 | 3099 (w) | 1606 (m) | 1043 (s) | 775 (s) | - | 341(4.63) |
| 3 | - | 1584 (m) | 1014 (s) | 750 (s) | 1.60 | 352(4.93); 601(2.57) |
| 4 | - | 1579 (m) | 1011 (s) | 732 (s) | Diamagnetic | 354(4.90); 609(2.08) |
| 5 | - | 1593 (m) | 947 (s) | 729 (s) | Diamagnetic | 354(5.21) |
| 6 | - | 1573 (m) | 964 (s) | 732 (s) | 1.67 | 624(2.23); 422(3.92); 324(4.61) |
| 7 | - | 1573 (m) | 969 (s) | 770 (s) | Diamagnetic | 342(4.88); 444(3.55); 609(2.07) |
| 8 | - | 1599 (m) | 957 (s) | 731 (s) | Diamagnetic | 341(5.08) |
2.4. Magnetic Susceptibility and Electronic Spectral Studies
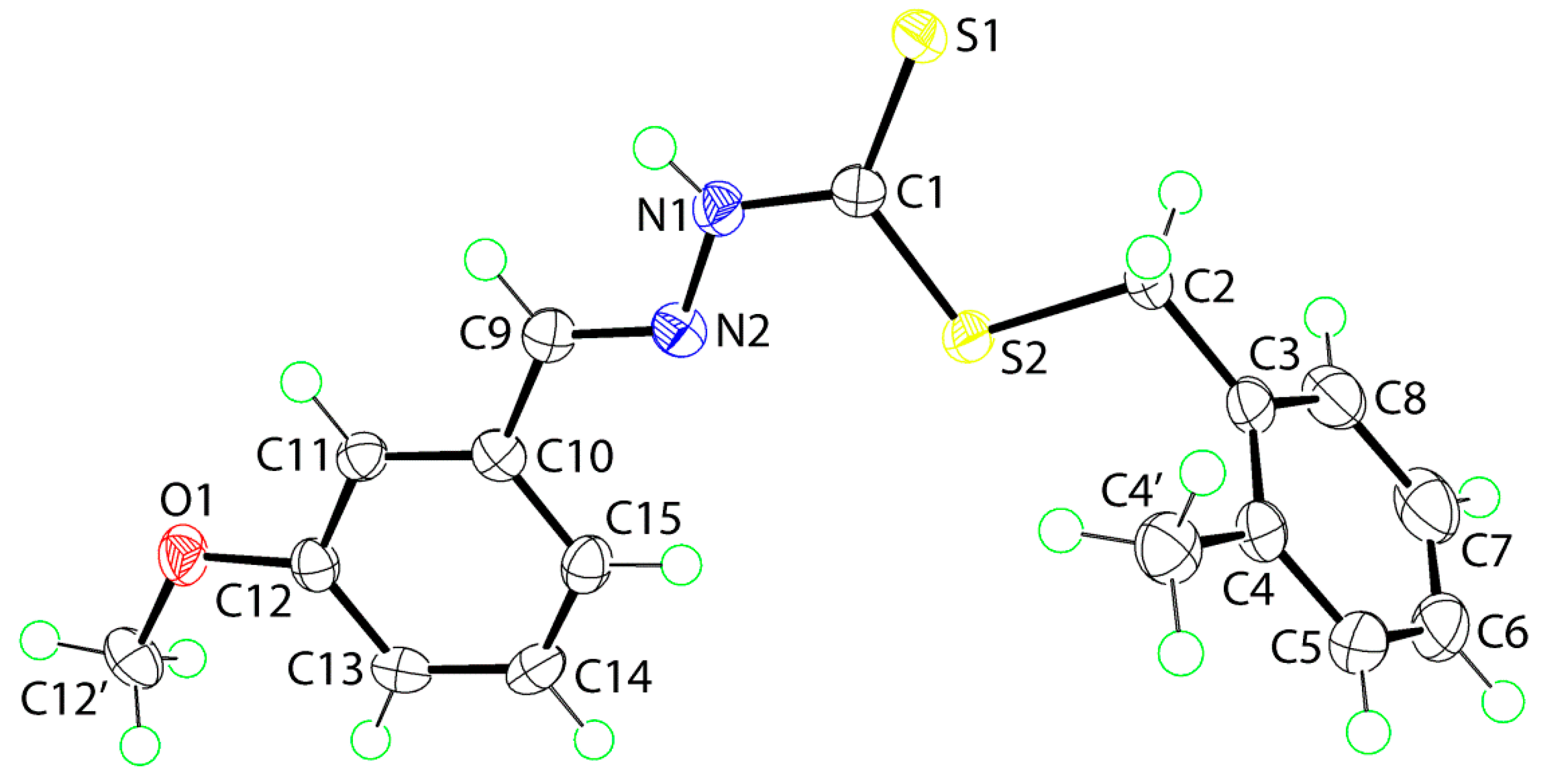
2.5. Crystal Data and Molecular Structures
2.5.1. Molecular Structures
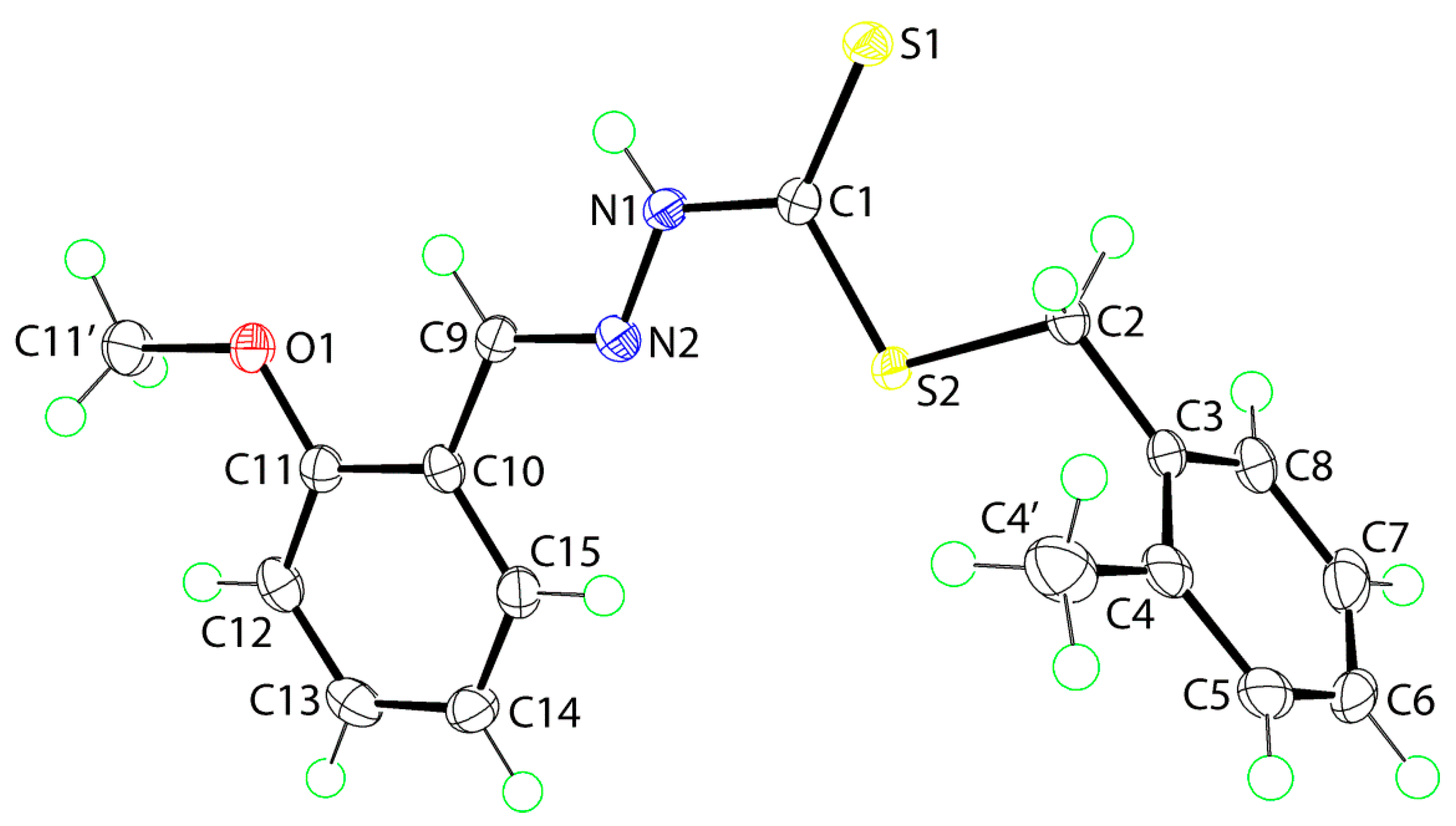
| Parameter | 1 | 2 |
|---|---|---|
| C1‒S1 | 1.6613(14) | 1.6724(14) |
| C1‒S2 | 1.7587(14) | 1.7537(14) |
| C2‒S2 | 1.8245(14) | 1.8241(15) |
| C1‒N1 | 1.3470(17) | 1.3373(19) |
| N1‒N2 | 1.3694(16) | 1.3765(17) |
| C9‒N2 | 1.2821(19) | 1.2797(19) |
| S1‒C1‒S2 | 124.77(8) | 124.68(9) |
| S1‒C1‒N1 | 122.02(10) | 121.68(11) |
| S2‒C1‒N1 | 113.21(10) | 113.64(11) |
| C1‒N1‒N2 | 118.76(11) | 119.64(12) |
| N1‒N2‒C9 | 116.56(12) | 115.74(12) |

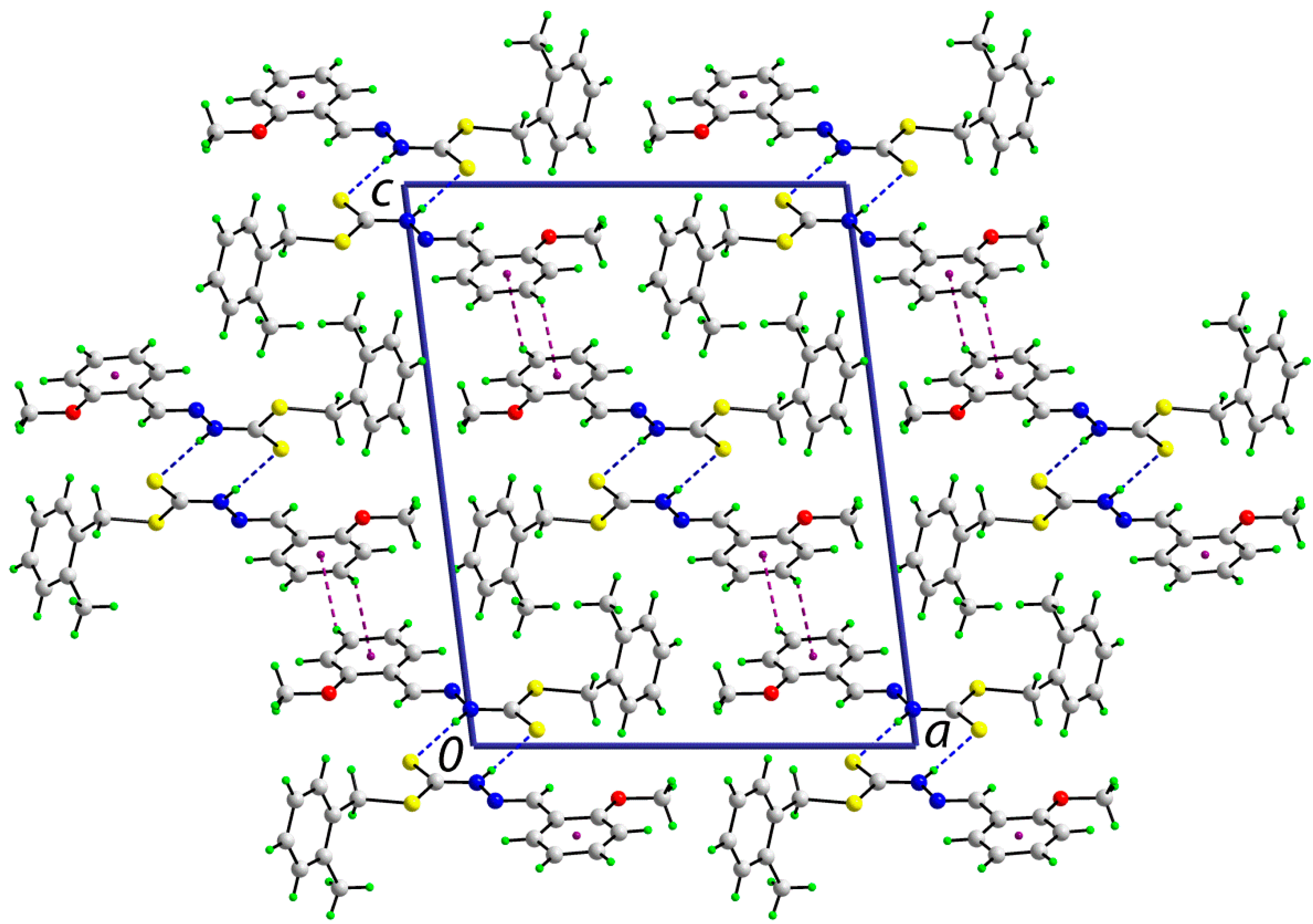
2.5.2. Supramolecular Architectures
| A | H | B | H···B (Å) | A···B(Å) | A-H-B(°) | Symmetry Operation |
|---|---|---|---|---|---|---|
| 1 | ||||||
| N1 | H1n | S1 | 2.503(14) | 3.3665(12) | 172.6(13) | 1-x, 3-y, 1-z |
| C13 | H13 | Cg(C10–C15) | 2.77 | 3.5374(15) | 139 | ½- x, -½+y, 1½-z |
| 2 | ||||||
| N1 | H1n | S1 | 2.53(2) | 3.3688(15) | 168.5(19) | -x, -y, -z |
| C11 | H11 | O1 | 2.44 | 3.351(2) | 160 | -x, -½+y, ½-z |
| C12' | H12b | S1 | 2.84 | 3.5574(17) | 130 | x, 1½-y, ½+z |
| C12' | H12c | Cg(C10–C15) | 2.80 | 3.6949(19) | 152 | x, 1½-y, ½+z |

2.6. Cytotoxic Activity
2.7. DNA Binding Studies
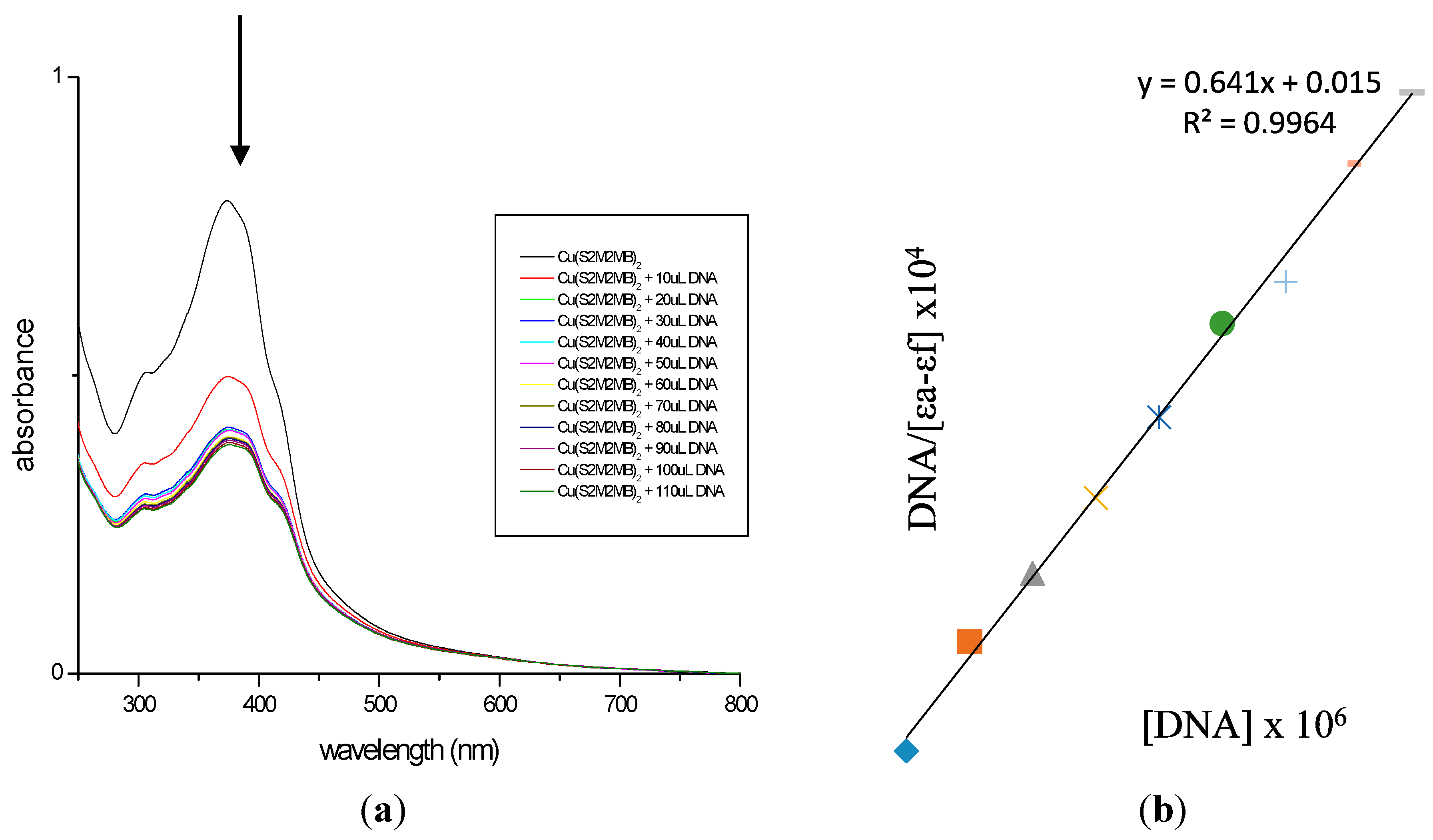
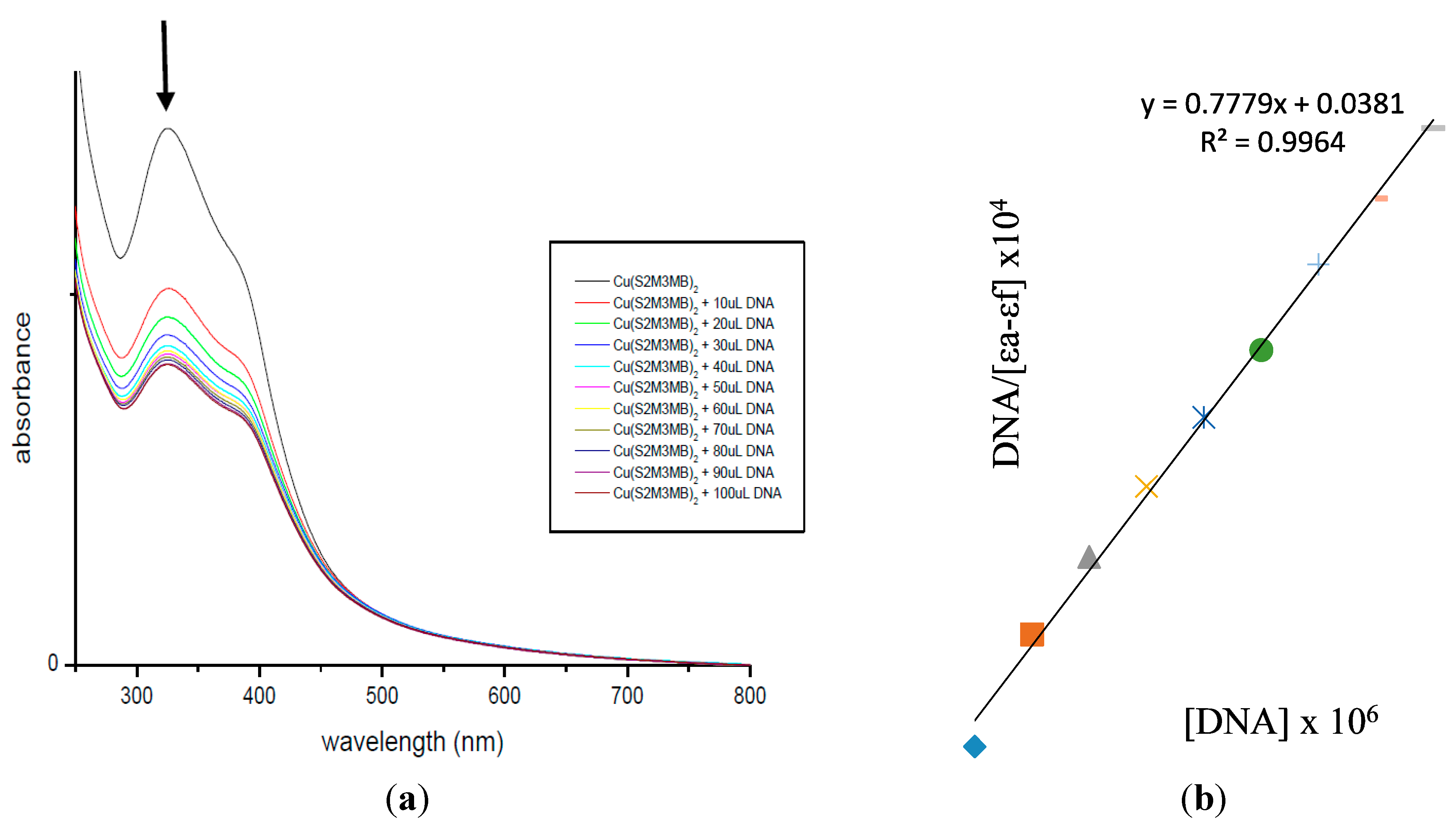
3. Experimental Section
3.1. Chemicals
3.2. Syntheses
3.2.1. S-2-Methybenzyldithiocarbazate (S2MBDTC)
3.2.2. S-2-Methylbenzyl-β-N-(2-benzyl-2-methoxymethylene)dithiocarbazate (NS1) (1)
3.2.3. S-2-Methylbenzyl-β-N-(2-benzyl-3-methoxymethylene)dithiocarbazate (NS2) (2)
3.2.4. Metal Complexes Derived from Ni(II) Acetate, Cu(II) Acetate, and Zn(II) Acetate
3.3. X-ray Crystallography
3.4. Cytotoxic Assay
| Compound | 1 | 2 |
|---|---|---|
| Formula | C17H18N2OS2 | C17H18N2OS2 |
| Formula weight | 330.45 | 330.45 |
| Crystal system | monoclinic | monoclinic |
| Space group | P21/n | P21/c |
| a/Å | 14.4313(2) | 17.6572(6) |
| b/Å | 6.0887(1) | 5.2747(2) |
| c/Å | 18.4462(3) | 19.3205(7) |
| β/° | 97.074(2) | 114.736(4) |
| V/Å3 | 1608.49(4) | 1634.33(11) |
| Z | 4 | 4 |
| Dc/g·cm−3 | 1.365 | 1.343 |
| F(000) | 696 | 696 |
| μ/mm−1 | 3.017 | 2.969 |
| Measured data | 19,873 | 10,986 |
| θ range/° | 3.7–71.4 | 4.6–71.4 |
| Unique data | 3128 | 3151 |
| Observed data (I ≥ 2.0σ(I)) | 3058 | 2965 |
| R, obs. data; all data | 0.033; 0.034 | 0.033; 0.035 |
| a, b in weighting scheme | 0.057, 0.763 | 0.052, 0.787 |
| Rw, obs. data; all data | 0.089; 0.089 | 0.086; 0.088 |
| Residual electron density peaks/e Å3 | 0.31, −0.42 | 0.43, −0.30 |
3.5. DNA Binding Studies
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Torre, M.H.; Gambino, D.; Araujo, J.; Ceretto, H.; González, M.; Lavanggi, M.L.; Azqueta, A.; Cerain, A.L.; Vega, A.M.; Abram, U.; et al. Novel Cu(II) quinoxaline N1,N4-dioxide complexes as selective hypoxic cytotoxins. Eur. J. Med. Chem. 2005, 40, 473–483. [Google Scholar] [CrossRef] [PubMed]
- Richardson, D.R.; Bernhardt, P.V. Crystal and molecular structure of 2-hydroxy-1-naphthaldehyde isonicotinoyl hydrazone (NIH) and its iron (III) complex: an iron chelator with anti-tumour activity. J. Biol. Inorg. Chem. 1999, 4, 266–273. [Google Scholar] [CrossRef] [PubMed]
- Blanco, A.G.; Sola, M.; Gomis-Rüth, F.X.; Coll, M. Tandem DNA recognition by PhoB, a two-component signal transduction transcriptional activator. Structure 2002, 10, 701–713. [Google Scholar] [CrossRef] [PubMed]
- Fukushima, T.; Takata, M.; Morrison, C.; Araki, R.; Fujimori, A.; Abe, M.; Takeda, S. Genetic analysis of the DNA-dependent protein kinase reveals an inhibitory role of Ku in late S–G2 phase DNA double-strand break repair. J. Biol. Chem. 2001, 276, 44413–44418. [Google Scholar] [CrossRef] [PubMed]
- Gomathi, V.; Selvameena, R. Synthesis, characterisation and biological studies of complexes of 3D transition metals and with Schiff base derived from sulfadiazine and 2-acetylnaphthalene. Int. J. Recent Sci. Res. 2013, 4, 94–97. [Google Scholar]
- Shivakumar, K.; Shashidhar, P.; Vithal Reddy, P.; Halli, M.B. Synthesis, spectral characterisation and biological activity of benzofuran Schiff bases with Co(II), Ni(II), Cu(II), Zn(II), Cd(II) and Hg(II) complexes. J. Coord. Chem. 2008, 61, 2274–2287. [Google Scholar] [CrossRef]
- Hunashal, R.D.; Satyanarayana, D. One pot synthesis of 3-(substituted phenoxymethyl)-6-phenyl/substituted phenoxymethyl-1,2,4-triazolo[3,4-b][1,3,4] thiadiazole derivatives as antimicrobial agents. Int. J. Pharm. Biol. Sci. 2012, 3, 183–192. [Google Scholar]
- Tarafder, M.T.H.; Ali, M.A.; Wee, D.J.; Azahari, K.; Silong, S.; Crowse, K.A. Complexes of a tridentate ONS Schiff base. Synthesis and biological properties. Transit. Metal Chem. 2000, 25, 456–460. [Google Scholar] [CrossRef]
- Dharamraj, N.; Viswanathanmurthi, P.; Natarajan, K. Ruthenium (II) complexes containing bidentate Schiff bases and their antifungal activity. Transit. Metal Chem. 2001, 26, 105–109. [Google Scholar] [CrossRef]
- Ejiah, F.N.; Fasina, T.M.; Familoni, O.B.; Ogunsola, F.T. Substituent effect on spectral and antimicrobial activity of Schiff bases derived from aminobenzoic acids. Adv. Biol. Chem. 2013, 3, 475–479. [Google Scholar] [CrossRef]
- Pedreño, E.; López-Contreras, A.J.; Cremades, A.; Peñafiel, R. Protecting or promoting effects of spermine on DNA strand breakage induced by iron or copper ions as a function of metal concentration. J. Inorg. Biochem. 2005, 99, 2074–2080. [Google Scholar] [CrossRef] [PubMed]
- Routier, S.; Vezin, H.; Lamour, E.; Bernier, J.L.; Catteau, J.P.; Bailly, C. DNA cleavage by hydroxy-salicylidene-ethylendiamine-iron complexes. Nucleic Acids Res. 1999, 27, 4160–4166. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.K.; Tripathi, P.; Bharty, M.K.; Srivastava, A.K.; Singh, S.; Butcher, R.J. Ni(II) and Mn(II) complexes of NNS tridentate ligand N′-[(2-methoxyphenyl)carbonothioyl] pyridine-2-carbohydrazide (H2mcph): Synthesis, spectral and structural characterization. Polyhedron 2010, 29, 1939–1945. [Google Scholar] [CrossRef]
- Walcourt, A.; Loyevsky, M.; Lovejoy, D.B.; Gordeuk, V.R.; Richardson, D. Novel aroylhydrazone and thiosemicarbazone iron chelators with antimalarial activity against chloroquine resistant and sensitive parasites. Int. J. Biochem. 2004, 36, 401–407. [Google Scholar]
- Livingstone, S.E.; Mihkelson, A.E. Metal chelates of biologically important compounds. II. Nickel complexes of dialkyldithiophosphates and their adducts with nitrogen heterocycles. Inorg. Chem. 1970, 9, 2545–2551. [Google Scholar] [CrossRef]
- Ali, M.A.; Livingstone, S.E. Metal complexes of sulphur-nitrogen chelating agents. Coord. Chem. Rev. 1974, 13, 101–132. [Google Scholar] [CrossRef]
- Subbaraj, P.; Ramu, A.; Raman, N.; Dharmaraja, J. Synthesis, characterization and pharmacological aspects of metal (II) complexes incorporating 4-[phenyl (phenylimino) methyl] benzene-1, 3-diol. J. Coord. Chem. 2014, 67, 2747–2764. [Google Scholar] [CrossRef]
- Spielmann, H.; Müller, L.; Averbeck, D.; Balls, M.; Brendler-Schwaab, S.; Castell, J.V.; Vohr, H.W. The second ECVAM workshop on phototoxicity testing. The report and recommendations of ECVAM workshop42. Altern. Lab. Anim. 2000, 28, 777–814. [Google Scholar] [PubMed]
- Tarafder, M.T.H.; Jin, K.T.; Crouse, K.A.; Ali, M.A.; Yamin, B.M.; Fun, H.-K. Coordination chemistry and bioactivity of Ni2+, Cu2+, Cd2+ and Zn2+ complexes containing bidentate Schiff bases derived from S-benzyldithiocarbazate and the X-ray crystal structure of bis[S-benzyl-β-N-(5-methyl-2-furylmethylene)dithiocarbazato]cadmium(II). Polyhedron 2002, 21, 2547–2554. [Google Scholar] [CrossRef]
- Dilović, I.; Rubčić, M.; Vrdoljak, V.; Pavelić, S.K.; Kralj, M.; Piantanida, I.; Cindrić, M. Novel thiosemicarbazone derivatives as potential antitumor agents: synthesis, physicochemical and structural properties, DNA interactions and antiproliferative activity. Bioorg. Med. Chem. 2008, 16, 5189–5198. [Google Scholar] [CrossRef] [PubMed]
- Mohan, M.; Sharma, P.; Jha, N.K. Some metal (II) chelates of 4-(m-aminophenyl)-2-formylpyridine thiosemicarbazone: Their preparation, characterizafion and antitumour activity. Inorg. Chim. Acta 1985, 107, 91–95. [Google Scholar] [CrossRef]
- Malhotra, R.; Singh, J.P.; Dudeja, M.; Dhindsa, K.S. Ligational behavior of N-substituted acid hydrazides towards transition metals and potentiation of their microbiocidal activity. J. Inorg. Biochem. 1992, 46, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.A.; Majumder, S.M.; Butcher, R.J.; Jasinski, J.P.; Jasinski, J.M. The preparation and characterization of bis-chelated nickel (II) complexes of the 6-methylpyridine-2-carboxaldehyde Schiff bases of S-alkyldithiocarbazates and the X-ray crystal structure of the bis {S-methyl-β-N-(6-methylpyrid-2-yl)-methylenedithiocarbazato} nickel(II) complex. Polyhedron 1997, 16, 2749–2754. [Google Scholar] [CrossRef]
- Ali, M.A.; Mirza, A.H.; Butcher, R.J. Synthesis and characterization of copper (II) complexes of the methylpyruvate Schiff base of S-methyldithiocarbazate (Hmpsme) and the X-crystal structures of Hmpsme and [Cu(mpsme)Cl]. Polyhedron 2001, 20, 1037–1043. [Google Scholar] [CrossRef]
- Ali, M.A.; Mirza, A.H.; Hamid, M.H.S.; Bujang, F.H.; Bernardt, P.V. The preparation and characterization of tin(IV) complexes of 2-quinolinecarboxaldehyde Schiff bases of S-methyl- and S-benzyldithiocarbazates and the X-ray crystal and molecular structures of the 2-quinolinecarboxaldehyde Schiff base of S-benzyldithiocarbazate (Hqaldsbz) and its tin(IV) complex [Sn(qaldsbz)I3]. Polyhedron 2004, 23, 2405–2412. [Google Scholar] [CrossRef]
- Ali, M.A.; Tarafder, M.T.H. Metal complexes of sulphur and nitrogen-containing ligands: Complexes of s-benzyldithiocarbazate and a schiff base formed by its condensation with pyridine-2-carboxaldehyde. J. Inorg. Nucl. Chem. 1977, 39, 1785–1791. [Google Scholar] [CrossRef]
- Tarafder, M.T.H.; Jin, K.T.; Crouse, K.A.; Ali, M.A.; Yamin, B.M.; Fun, H.-K. Coordination chemistry and bioactivity of some metal complexes containing two isomeric bidentate NS Schiff bases derived from S-benzyldithiocarbazate and the X-ray crystal structures of S-benzyl-β-N-(5-methyl-2-furylmethylene)dithiocarbazate and bis[S-benzyl-β-N-(2-furylmethylketone) dithiocarbazato]cadmium(II). Polyhedron 2002, 21, 2691–2698. [Google Scholar] [CrossRef]
- Ali, M.A.; Mirza, A.H.; Fereday, R.J.; Butcher, R.J.; Fuller, J.M.; Drew, S.C.; Gahan, L.R.; Hanson, G.R.; Moubaraki, B.; Murray, K.S. Synthetic, EPR spectroscopic, magnetic and X-ray crystallographic structural studies on copper (II) complexes of the tridentate NS donor ligand formed from 6-methyl-2-formylpyridine and S-methyldithiocarbazate (Hmpsme). Inorg. Chim. Acta 2005, 358, 3937–3948. [Google Scholar] [CrossRef]
- Tarafder, M.T.H.; Kasbollah, A.; Crouse, K.A.; Ali, A.M.; Yamin, B.M.; Fun, H.K. Synthesis and characterization of Zn(II) and Cd(II) complexes of S-benzyl-β-N-(2-pyridyl)methylene dithiocarbazate (HNNS): bioactivity of the HNNS Schiff base and its Zn(II), Cu(II) and Cd(II) complexes and the X-ray structure of the [Zn(NNS)2] complex. Polyhedron 2001, 20, 2363–2370. [Google Scholar] [CrossRef]
- Shan, S.; Tian, Y.-L.; Wang, S.-H.; Wang, W.-L.; Xu, Y.-L. Benzyl 3-[(E)-benzylidene] dithiocarbazate. Acta Crystallogr. 2008, 64. [Google Scholar] [CrossRef]
- Spek, A.L. Single-crystal structure validation with the program PLATON. J. Appl. Crystallogr. 2003, 36, 7–13. [Google Scholar] [CrossRef]
- Todorović, T.R.; Bacchi, A.; Juranić, N.O.; Sladić, D.M.; Pelizzi, G.; Božić, T.T.; Anđelković, K.K. Synthesis and characterization of novel Cd (II), Zn (II) and Ni (II) complexes with 2-quinolinecarboxaldehyde selenosemicarbazone. Crystal structure of bis (2-quinolinecarboxaldehyde selenosemicarbazonato) nickel (II). Polyhedron 2007, 26, 3428–3436. [Google Scholar] [CrossRef]
- Bharti, N.; Athar, F.; Maurya, M.R.; Azam, A. Synthesis, characterization and in vitro anti-amoebic activity of new palladium (II) complexes with 5-nitrothiophene-2-carboxaldehyde N (4)-substituted thiosemicarbazones. Bioorg. Med. Chem. 2004, 12, 4679–4684. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.P.; Singh, P.; Singh, A.K. Synthesis, structural and corrosion inhibition studies on cobalt (II), nickel (II), copper (II) and zinc (II) complexes with 2-acetylthiophene benzoylhydrazone. Inorg. Chim. Acta 2011, 379, 56–63. [Google Scholar] [CrossRef]
- Singh, K.; Barwa, M.S.; Tyagi, P. Synthesis and characterization of cobalt (II), nickel (II), copper (II) and zinc (II) complexes with Schiff base derived from 4-amino-3-mercapto-6-methyl-5-oxo-1, 2,4-triazine. Eur. J. Med. Chem. 2007, 42, 394–402. [Google Scholar] [CrossRef] [PubMed]
- Barton, J.K.; Danishefsky, A.; Goldberg, J. Tris (phenanthroline) ruthenium (II): Stereoselectivity in binding to DNA. J. Am. Chem. Soc. 1984, 106, 2172–2176. [Google Scholar] [CrossRef]
- Tysoe, S.A.; Morgan, R.J.; Baker, A.D.; Strekas, T.C. Spectroscopic investigation of differential binding modes of Δ and Δ-Ru(bpy)2(ppz)2+ with calf thymus DNA. J. Phys. Chem. 1993, 97, 1707–1711. [Google Scholar] [CrossRef]
- Kelly, J.M.; Tossi, A.B.; McConnell, D.J.; OhUigin, C. A study of the interactions of some polypyridylruthenium (II) complexes with DNA using fluorescence spectroscopy, topoisomerisation and thermal denaturation. Nucleic Acids Res. 1985, 13, 6017–6034. [Google Scholar] [CrossRef] [PubMed]
- Sangeetha Gowda, K.R.; Bhojya Naik, H.S.; Vinay Kumar, B.; Sudhamani, C.N.; Sudeep, H.V.; Ravikumar Naik, T.R.; Krishnamurthy, G. Synthesis, antimicrobial, DNA-binding and photonuclease studies of Cobalt(III) and Nickel(II) Schiff base complexes, Spectrochim. Acta Mol. Biomol. Spectrosc. 2013, 105, 229–237. [Google Scholar] [CrossRef]
- Wu, B.Y.; Gao, L.H.; Duan, Z.M.; Wang, K.Z. Syntheses and DNA-binding studies of two ruthenium(II) complexes containing one ancillary ligand of bpy or phen: [Ru(bpy)(pp[2,3]p)2](ClO4)2 and [Ru(phen)(pp[2,3]p)2](ClO4)2. J. Inorg. Biochem. 2005, 99, 1685–1691. [Google Scholar] [CrossRef] [PubMed]
- Long, E.C.; Barton, J.K. On demonstrating DNA intercalation. Acc. Chem. Res. 1990, 23, 271–273. [Google Scholar] [CrossRef]
- Kovala-Demertzi, D.; Miller, J.R.; Kourkoumelis, N.; Hadjikakou, S.K.; Demertzis, M.A. Palladium(II) and platinum(II) complexes of pyridine-2-carbaldehyde thiosemicarbazone with potential biological activity. Synthesis, structure and spectral properties. Extended network via hydrogen bond linkages of [Pd(PyTsc)Cl]. Polyhedron 1999, 18, 1005–1013. [Google Scholar] [CrossRef]
- Ravoof, T.B.S.A.; Crouse, K.A.; Tahir, M.I.M.; How, F.N.F.; Rosli, R.; Watkins, D. Synthesis, characterisation and biological activities of 2-methylbenzyl 2-(dipyridin-2-yl methylene)hydrazinecarbodithioate. J. Chem. Crystallogr. 2011, 41, 491–495. [Google Scholar] [CrossRef]
- CrysAlis, P.R.O. Agilent Technologies; Yarnton: Oxfordshire, UK, 2011. [Google Scholar]
- Sheldrick, G.M. A short history of SHELX. Acta Crystallogr. Sect. A 2008, 64, 112–122. [Google Scholar] [CrossRef]
- Farrugia, L.J. WinGX and ORTEP for Windows: An update. J. Appl. Crystallogr. 2012, 45, 849–854. [Google Scholar] [CrossRef]
- Gans, J.; Shalloway, D. Qmol: A program for molecular visualization on Windows-based PCs. J. Mol. Graph. Model. 2001, 19, 557–559. [Google Scholar] [CrossRef] [PubMed]
- Brandenburg, K. Diamond; Crystal Impact GbR: Bonn, Germany, 2006. [Google Scholar]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Badiger, D.S.; Hunoor, R.S.; Patil, B.R.; Vadavi, R.S.; Mangannavar, C.V.; Muchchandi, I.S.; Gudasi, K.B. Synthesis, spectroscopic properties and biological evaluation of transition metal complexes of salicylhydrazone of anthranilhydrazide: X-ray crystal structure of copper complex. Inorg. Chim. Acta 2012, 384, 197–203. [Google Scholar] [CrossRef]
- Reichmann, M.E.; Rice, S.A.; Thomas, C.A.; Doty, P. A further examination of the molecular weight and size of desoxypentose nucleic acid. J. Am. Chem. Soc. 1954, 76, 3047–3053. [Google Scholar] [CrossRef]
- Chaires, J.B.; Dattagupta, N.; Crothers, D.M. Studies on interaction of anthracycline antibiotics and deoxyribonucleic acid: Equilibrium binding studies on the interaction of daunomycin with deoxyribonucleic acid. Biochemistry 1982, 21, 3933–3940. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Md Yusof, E.N.; Ravoof, T.B.S.A.; Tiekink, E.R.T.; Veerakumarasivam, A.; Crouse, K.A.; Mohamed Tahir, M.I.; Ahmad, H. Synthesis, Characterization and Biological Evaluation of Transition Metal Complexes Derived from N, S Bidentate Ligands. Int. J. Mol. Sci. 2015, 16, 11034-11054. https://doi.org/10.3390/ijms160511034
Md Yusof EN, Ravoof TBSA, Tiekink ERT, Veerakumarasivam A, Crouse KA, Mohamed Tahir MI, Ahmad H. Synthesis, Characterization and Biological Evaluation of Transition Metal Complexes Derived from N, S Bidentate Ligands. International Journal of Molecular Sciences. 2015; 16(5):11034-11054. https://doi.org/10.3390/ijms160511034
Chicago/Turabian StyleMd Yusof, Enis Nadia, Thahira Begum S. A. Ravoof, Edward R. T. Tiekink, Abhimanyu Veerakumarasivam, Karen Anne Crouse, Mohamed Ibrahim Mohamed Tahir, and Haslina Ahmad. 2015. "Synthesis, Characterization and Biological Evaluation of Transition Metal Complexes Derived from N, S Bidentate Ligands" International Journal of Molecular Sciences 16, no. 5: 11034-11054. https://doi.org/10.3390/ijms160511034