Prediction of Protein–Protein Interactions with Clustered Amino Acids and Weighted Sparse Representation
Abstract
:1. Introduction
2. Results and Discussion
2.1. Evaluation Criteria
2.2. Assessment of Prediction Ability
| Cluster Profiles | Protein Blocks Method |
|---|---|
| CP(13) | G-IV-FYW-A-L-M-E-QRK-P-ND-HS-T-C |
| CP(11) | G-IV-FYW-A-LM-EQRK-P-ND-HS-T-C |
| CP(9) | G-IV-FYW-ALM-EQRK-P-ND-HS-TC |
| CP(8) | G-IV-FYW-ALM-EQRK-P-ND-HSTC |
| CP(5) | G-IVFYW-ALMEQRK-P-NDHSTC |
| n-Peptide | CP(13) | CP(11) | CP(9) | CP(8) | CP(5) |
|---|---|---|---|---|---|
| n = 1 | 13 | 11 | 9 | 8 | 5 |
| n = 2 | 169 | 121 | 81 | 64 | 25 |
| n = 3 | 2197 | 1331 | 729 | 512 | 125 |
| Criteria | CP(5) | CP(8) | CP(9) | CP(11) | CP(13) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dimension | 5 | 25 | 125 | 8 | 64 | 512 | 9 | 81 | 729 | 11 | 121 | 1331 | 13 | 169 | 2197 |
| ACC (%) | 71.16 | 82.85 | 87.43 | 77.90 | 87.74 | 90.91 | 79.63 | 88.19 | 90.86 | 81.47 | 89.05 | 90.77 | 82.99 | 89.46 | 90.26 |
| SN (%) | 71.26 | 84.21 | 89.88 | 78.02 | 90.02 | 94.17 | 79.92 | 90.72 | 94.15 | 81.78 | 91.52 | 94.17 | 83.75 | 92.23 | 93.32 |
| PE (%) | 70.94 | 80.86 | 84.38 | 77.70 | 84.90 | 87.22 | 79.14 | 85.10 | 87.13 | 80.99 | 86.08 | 86.91 | 81.88 | 86.19 | 86.71 |
| MCC (%) | 58.96 | 71.55 | 77.98 | 65.57 | 78.45 | 83.43 | 67.56 | 79.14 | 83.34 | 69.81 | 80.47 | 83.19 | 71.76 | 81.11 | 82.37 |
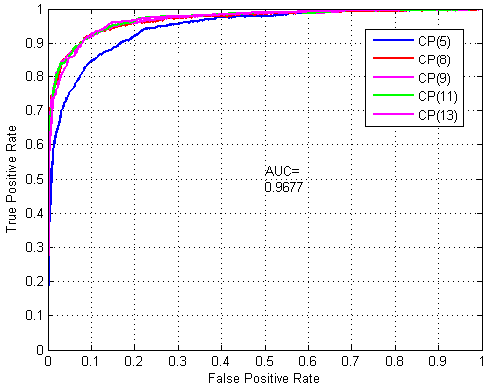
2.3. Robustness Performance Evaluation
| Criteria | CP(5) | CP(8) | CP(9) | CP(11) | CP(13) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dimension | 5 | 25 | 125 | 8 | 64 | 512 | 9 | 81 | 729 | 11 | 121 | 1331 | 13 | 169 | 2197 |
| ACC (%) | 66.63 | 76.34 | 81.43 | 72.89 | 81.68 | 81.90 | 73.19 | 81.79 | 80.81 | 74.43 | 82.60 | 78.50 | 76.19 | 83.04 | 77.29 |
| SN (%) | 64.76 | 72.45 | 77.67 | 70.67 | 77.90 | 77.56 | 70.44 | 78.39 | 76.27 | 71.29 | 78.77 | 73.41 | 72.61 | 79.20 | 71.69 |
| PE (%) | 72.65 | 85.12 | 88.47 | 78.18 | 88.51 | 89.94 | 79.89 | 88.03 | 89.49 | 81.67 | 89.41 | 89.72 | 84.03 | 89.75 | 90.37 |
| MCC (%) | 55.14 | 63.29 | 69.49 | 60.28 | 69.75 | 70.02 | 60.40 | 69.96 | 68.53 | 61.56 | 71.00 | 65.38 | 63.29 | 71.60 | 63.73 |
2.4. Comparison with Other Methods
| Criteria | Auto Covariance | Conjoint Triad | Moran Autocorrelation | Geary Autocorrelation | Pseudo-Amino Acid Composition | Our Method |
|---|---|---|---|---|---|---|
| ACC (%) | 88.89 | 89.57 | 88.90 | 85.06 | 86.45 | 90.91 |
| SN (%) | 92.07 | 92.91 | 92.12 | 88.20 | 88.16 | 94.17 |
| PE (%) | 85.12 | 85.68 | 85.08 | 80.97 | 84.23 | 87.22 |
| MCC (%) | 80.19 | 81.26 | 80.21 | 74.51 | 76.56 | 84.43 |
| Criteria | CP(5) | CP(8) | CP(9) | CP(11) | CP(13) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dimension | 5 | 25 | 125 | 8 | 64 | 512 | 9 | 81 | 729 | 11 | 121 | 1331 | 13 | 169 | 2197 |
| ACC (%) | 67.27 | 74.80 | 81.44 | 72.05 | 82.81 | 88.17 | 72.86 | 83.32 | 89.23 | 74.58 | 84.89 | 90.81 | 75.66 | 86.27 | 92.04 |
| SN (%) | 71.04 | 75.56 | 80.49 | 75.20 | 82.64 | 86.35 | 75.62 | 83.18 | 87.48 | 77.20 | 84.45 | 88.85 | 77.60 | 85.46 | 90.37 |
| PE (%) | 66.06 | 74.44 | 82.06 | 70.76 | 82.95 | 89.61 | 71.69 | 83.43 | 90.66 | 73.36 | 85.22 | 92.48 | 74.71 | 86.88 | 93.51 |
| MCC (%) | 55.84 | 62.29 | 69.78 | 59.64 | 71.53 | 79.12 | 60.39 | 72.21 | 80.76 | 62.03 | 74.34 | 83.30 | 63.14 | 76.31 | 85.35 |
| Criteria | WSRC | SRC | SVM | Guo’s Work | Zhou’s Work | Yang’s Work |
|---|---|---|---|---|---|---|
| ACC (%) | 90.91 | 87.46 | 88.17 | 89.33 | 88.56 | 86.15 |
| SN (%) | 94.17 | 89.93 | 86.55 | 89.93 | 87.37 | 81.03 |
| PE (%) | 87.22 | 84.39 | 89.61 | 88.87 | 89.50 | 90.24 |
| MCC (%) | 83.43 | 78.02 | 79.12 | N/A | 77.15 | N/A |
3. Experimental Section
3.1. Dataset
3.2. Pseudo-Amino Acid Composition and Reduced Amino Acid Alphabet
3.3. Weighted Sparse Representation Based Classification
s.t. y = Xα
s.t. y = Xα
s.t. ‖y − Xα‖ ≤ ε
s.t. ‖y − Xα‖ ≤ ε
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Schelhorn, S.E.; Mestre, J.; Albrecht, M.; Zotenko, E. Inferring physical protein contacts from large-scale purification data of protein complexes. Mol. Cell. Proteomics 2011, 10, 2889–2894. [Google Scholar] [CrossRef]
- Pawson, T.; Nash, P. Protein–protein interactions define specificity in signal transduction. Genes Dev. 2000, 14, 1027–1047. [Google Scholar] [PubMed]
- MacPherson, R.E.K.; Ramos, S.V.; Vandenboom, R.; Roy, B.D.; Peters, S.J. Skeletal muscle PLIN proteins, ATGL and CGI-58, interactions at rest and following stimulated contraction. Am. J. Physiol. 2013, 304, 644–650. [Google Scholar]
- Phizicky, E.; Fields, S. Protein–protein interactions: Methods for detection and analysis. Microbiol. Rev. 1995, 59, 94–123. [Google Scholar] [PubMed]
- Young, K.H. Yeast two-hybrid: So many interactions, (in) so little time. Biol. Reprod. 1998, 58, 302–311. [Google Scholar] [CrossRef] [PubMed]
- Puig, O.; Caspary, F.; Rigaut, G.; Rutz, B.; Bouveret, E.; Bragado-Nilsson, E.; Wilm, M.; Seraphin, B. The tandem affinity purification (TAP) method: A general procedure of protein complex purification. Methods (San Diego, Calif.) 2001, 24, 218–229. [Google Scholar] [CrossRef]
- Lei, Y.K.; You, Z.H.; Dong, T.; Jiang, Y.X.; Yang, J.A. Increasing reliability of protein interactome by fast manifold embedding. Pattern Recognit. Lett. 2013, 34, 372–379. [Google Scholar] [CrossRef]
- Zhu, L.; You, Z.H.; Huang, D.S.; Wang, B. t-LSE: A novel robust geometric approach for modeling protein–protein interaction networks. PLoS ONE 2013, 8, e58368. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.K.; You, Z.H.; Ji, Z.; Zhu, L.; Huang, D.S. Assessing and predicting protein interactions by combining manifold embedding with multiple information integration. BMC Bioinform. 2012, 13, S3. [Google Scholar] [CrossRef]
- You, Z.H.; Lei, Y.K.; Gui, J.; Huang, D.S.; Zhou, X. Using manifold embedding for assessing and predicting protein interactions from high-throughput experimental data. Bioinformatics 2010, 26, 2744–2751. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; You, Z.H.; Zhou, M.C.; Li, S.; Leung, H.; Xia, Y.N.; Zhu, Q.S. A highly efficient approach to protein interactome mapping based on collaborative filtering framework. Scientific Rep. 2015, 5. [Google Scholar] [CrossRef]
- Rao, V.S.; Srinivas, K.; Sujini, G.N.; Kumar, G.N.S. Protein–protein interaction detection: Methods and analysis. Int. J. Proteomics 2014, 2014, 147648. [Google Scholar] [CrossRef] [PubMed]
- Hosur, R.; Xu, J.; Bienkowska, J.; Berger, B. iWRAP: An interface threading approach with application to prediction of cancer-ralated protein–protein interactions. J. Mol. Biol. 2011, 405, 1295–1310. [Google Scholar] [CrossRef] [PubMed]
- Valente, G.T.; Acencio, M.L.; Martins, C.; Lemke, N. The development of a universal in silico predictor of protein–protein interactions. PLoS ONE 2013, 8, e65587. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zhang, D.; Xu, R.; Xu, J.; Wang, X.; Chen, Q.; Dong, Q.; Chou, K.C. Combining evolutionary information extracted from frequency profiles with sequence-based kernels for protein remote homology detection. Bioinformatics 2014, 30, 472–479. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Wang, X.; Zou, Q.; Dong, Q.; Chen, Q. Protein remote homology detection by combining Chou’s pseudo zmino zcid composition and profile-based protein representation. Mol. Inform. 2013, 32, 775–782. [Google Scholar] [CrossRef]
- Liu, B.; Xu, J.; Zou, Q.; Xu, R.; Wang, X.; Chen, Q. Using distances between Top-n-gram and residue pairs for protein remote homology detection. BMC Bioinform. 2014, 15, 133–139. [Google Scholar] [CrossRef]
- Liu, B.; Xu, J.; Fan, S.; Xu, R.; Zhou, J.; Wang, X. PseDNA-Pro: DNA-binding protein identification by combining Chou’s PseAAC and physicochemical distance transformation. Mol. Inform. 2015, 34, 8–17. [Google Scholar] [CrossRef]
- Shen, J.; Zhang, J.; Luo, X.; Zhu, W.; Yu, K.; Chen, K.; Li, Y.; Jiang, H. Predicting protein–protein interactions based only on sequences information. Proc. Natl. Acad. Sci. USA 2007, 104, 4337–4341. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Yu, L.; Wen, Z.; Li, M. Using support vector machine combined with auto covariance to predict protein–protein interactions from protein sequences. Nucleic Acids Res. 2008, 36, 3025–3030. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.C. Prediction of protein cellular attributes using pseudo-amino acid composition. Proteins 2001, 44, 60–60. [Google Scholar] [CrossRef]
- Mohabatkar, H.; Beigi, M.M.; Esmaeili, A. Prediction of GABAA receptor proteins using the concept of Chou’s pseudo-amino acid composition and support vector machine. J. Theor. Biol. 2011, 281, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Rao, N.; Liu, G.; Yang, Y.; Wang, G. Predicting protein folding rates using the concept of Chou’s pseudo amino acid composition. J. Comput. Chem. 2011, 32, 1612–1617. [Google Scholar] [CrossRef] [PubMed]
- Zou, D.; He, Z.; He, J.; Xia, Y. Supersecondary structure prediction using Chou’s pseudo amino acid composition. J. Comput. Chem. 2011, 32, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.W.; Zhang, Y.L.; Yang, H.F.; Zhao, C.H.; Pan, Q. Using the concept of Chou’s pseudo amino acid composition to predict protein subcellular localization: An approach by incorporating evolutionary information and von Neumann entropies. Amino Acids 2008, 34, 565–572. [Google Scholar] [CrossRef] [PubMed]
- Mei, S. Predicting plant protein subcellular multi-localization by Chou’s PseAAC formulation based multi-label homolog knowledge transfer learning. J. Theor. Biol. 2012, 310, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Diaz, H.; Gonzalez-Diaz, Y.; Santana, L.; Ubeira, F.M.; Uriarte, E. Proteomics, networks and connectivity indices. Proteomics 2008, 8, 750–778. [Google Scholar] [CrossRef] [PubMed]
- Du, P.; Wang, X.; Xu, C.; Gao, Y. PseAAC-Builder: A cross-platform stand-alone program for generating various special Chou’s pseudo-amino acid compositions. Anal. Biochem. 2012, 425, 117–119. [Google Scholar] [CrossRef] [PubMed]
- Cao, D.S.; Xu, Q.S.; Liang, Y.Z. Propy: A tool to generate various modes of Chou’s PseAAC. Bioinformatics 2013, 29, 960–962. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Liu, F.; Fang, L.; Wang, X.; Chou, K.C. repDNA: A Python package to generate various modes of feature vectors for DNA sequences by incorporating user-defined physicochemical properties and sequence-order effects. Bioinformatics 2015, 31, 1307–1309. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.D.; Dill, K.A. An iterative method for extracting energy-like quantities from protein structures. Proc. Natl. Acad. Sci. USA 1996, 93, 11628–11633. [Google Scholar] [CrossRef] [PubMed]
- Mirny, L.A.; Shakhnovich, E.I. Universally conserved positions in protein folds: Reading evolutionary signals about stability, folding kinetics and function. J. Mol. Biol. 1999, 291, 177–196. [Google Scholar] [CrossRef] [PubMed]
- Solis, A.D.; Rackovsky, S. Optimized representations and maximal information in proteins. Proteins 2000, 38, 149–164. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Fang, L.; Chen, J.; Liu, F.; Wang, X. miRNA-dis: MicroRNA precursor identification based on distance structure status pairs. Mol. BioSyst. 2015, 11, 1194–1204. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Fang, L.; Liu, F.; Wang, X. iMiRNA-PseDPC: MicroRNA precursor identification with a pseudo distance-pair composition approach. J. Biomol. Struct. Dyn. 2015. [Google Scholar] [CrossRef]
- Wright, J.; Yang, A.; Ganesh, A.; Sastry, S.; Ma, Y. Robust face recognition via sparse representation. Pattern Anal. Mach. Intell. IEEE Trans. 2009, 31, 210–227. [Google Scholar] [CrossRef]
- Lu, C.Y.; Min, H.; Gui, J.; Zhu, L.; Lei, Y.K. Face recognition via weighted sparse representation. J. Vis. Commun. Image Represent. 2013, 24, 111–116. [Google Scholar] [CrossRef]
- Zhou, Y.Z.; Gao, Y.; Zheng, Y.Y. Prediction of protein–protein interactions using local description of amino acid sequence. Commun. Comput. Inf. Sci. 2011, 202, 254–262. [Google Scholar]
- Yang, L.; Xia, J.; Gui, J. Prediction of protein–protein interactions from protein sequence using local descriptors. Protein Pept. Lett. 2010, 17, 1085–1090. [Google Scholar] [CrossRef] [PubMed]
- Wootton, J.C.; Federhen, S. Statistics of local complexity in amino acid sequences and sequence databases. Comput. Chem. 1993, 17, 149–163. [Google Scholar] [CrossRef]
- Khosravian, M.; Faramarzi, F.K.; Beigi, M.M.; Behbahani, M. Predicting antibacterial peptides by the concept of Chou’s pseudo-amino acid composition and machine learning methods. Protein Pept. Lett. 2013, 20, 180–186. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Ding, H. Predicting ion channels and their types by the dipeptide mode of pseudo amino acid composition. J. Theor. Biol. 2011, 269, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Li, Q.Z. Using pseudo amino acid composition to predict protein structural class: Approached by incorporating 400 dipeptide components. J. Comput. Chem. 2007, 28, 1463–1466. [Google Scholar] [CrossRef] [PubMed]
- Liao, B.; Jiang, J.B.; Zeng, Q.G.; Zhu, W. Predicting apoptosis protein subcellular location with PseAAC by incorporating tripeptide composition. Protein Pept. Lett. 2011, 18, 1086–1092. [Google Scholar] [CrossRef] [PubMed]
- De Brevern, A.G.; Etchebest, C.; Hazout, S. Bayesian probabilistic approach for predicting backbone structures in terms of protein blocks. Proteins 2000, 41, 271–287. [Google Scholar] [CrossRef] [PubMed]
- Joseph, A.P.; Agarwal, G.; Mahajan, S.; Gelly, J.C.; Swapna, L.S.; Offmann, B.; Cadet, F.; Bornot, A.; Tyagi, M.; Valadié, H.; et al. A short survey onprotein blocks. Biophys. Rev. 2010, 2, 137–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, P.M.; Chen, W.; Lin, H.; Chou, K.C. iHSP-PseRAAAC: Identifying the heat shock protein families using pseudo reduced amino acid alphabet composition. Anal. Biochem. 2013, 442, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Feng, P.; Lin, H.; Chen, W.; Zuo, Y.C. Predicting the types of J-proteins using clustered amino acids. BioMed. Res. Int. 2014, 2014, 935719. [Google Scholar] [PubMed]
- Liu, B.; Xu, J.; Lan, X.; Xu, R.; Zhou, J.; Wang, X.; Chou, K.C.; Liu, B.; Xu, J.; Lan, X. iDNA-Prot|dis: Identifying DNA-binding proteins by incorporating amino acid distance-pairs and reduced alphabet profile into the general pseudo amino acid composition. PLoS ONE 2014, 9, e106691. [Google Scholar] [CrossRef] [PubMed]
- Etchebest, C.; Benros, C.; Bornot, A.; Camproux, A.C.; de Brevern, A. A reduced amino acid alphabet for understanding and designing protein adaptation to mutation. Eur. Biophys. J. 2007, 36, 1059–1069. [Google Scholar] [CrossRef] [PubMed]
- Vavasis, S.A. Nonlinear Optimization: Complexity Issues; Oxford University Press, Inc.: New York, NY, USA, 1991. [Google Scholar]
- Candes, E.J. The restricted isometry property and its implications for compressed sensing. Comptes Rendus Math. 2008, 346, 589–592. [Google Scholar] [CrossRef]
- Chen, S.S.; Donoho, D.L.; Michael.; Saunders, A. Atomic decomposition by basis pursuit. SIAM J. Sci. Comput. 1998, 20, 33–61. [Google Scholar] [CrossRef]
- Georgiou, D.N.; Karakasidis, T.E.; Nieto, J.J.; Torres, A. A study of entropy/clarity of genetic sequences using metric spaces and fuzzy sets. J. Theor. Biol. 2010, 267, 95–105. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Q.; You, Z.; Zhang, X.; Zhou, Y. Prediction of Protein–Protein Interactions with Clustered Amino Acids and Weighted Sparse Representation. Int. J. Mol. Sci. 2015, 16, 10855-10869. https://doi.org/10.3390/ijms160510855
Huang Q, You Z, Zhang X, Zhou Y. Prediction of Protein–Protein Interactions with Clustered Amino Acids and Weighted Sparse Representation. International Journal of Molecular Sciences. 2015; 16(5):10855-10869. https://doi.org/10.3390/ijms160510855
Chicago/Turabian StyleHuang, Qiaoying, Zhuhong You, Xiaofeng Zhang, and Yong Zhou. 2015. "Prediction of Protein–Protein Interactions with Clustered Amino Acids and Weighted Sparse Representation" International Journal of Molecular Sciences 16, no. 5: 10855-10869. https://doi.org/10.3390/ijms160510855