1. Introduction
MicroRNAs (miRNAs) are approximately 22-nt non-coding RNAs and they act as negative regulators of gene expression either by inhibiting mRNA translation or promoting mRNA degradation through partial base pairing at the 3'-untranslated region (3'-UTR) of the target mRNAs [
1,
2]. miRNAs play a key role in diverse biological processes such as organ development, cell proliferation, tumorigenesis, fat metabolism, behavior and embryogenesis [
3,
4]. Furthermore, abnormal expression of miRNAs may result in disease, dramatic phenotype variation, and even death [
5]. It is apparent that assays of expression patterns of miRNAs in development, disease and other cellular processes exhibit both theoretical and practical significances. However, the crucial issue for the assay is to select suitable reference genes for accurate calibration.
Several modern technologies, such as microarrays, high-throughput sequencing, bead-based flow-cytometry, northern blot and RT-qPCR have been used for determination and quantification of miRNA expression [
6,
7,
8]. The advantages of RT-qPCR for gene expression analysis are sensitivity and relatively accurate, wide dynamic range, relatively low-cost and low template requirements [
9]. Different strategies have been used to normalize RT-qPCR data [
10,
11,
12], and selection of reliable reference genes as internal controls has to be carefully considered before any experimental design [
13]. However, certain criteria have to be fulfilled for reference gene selection [
12,
14]. The stability of a given reference gene should be experimentally validated in each study. miRNA data normalization has usually relied on some other small RNA expression analyses, such as small nuclear (snRNAs) or small nucleolus (snoRNAs) RNAs (e.g., U8, U6 or RNU44). However, there are some potential disadvantages in stability for these references as they are produced and/or processed by different pathways [
12]. Recently, a few studies have explored the stability of some miRNAs in vertebrate species. Davoren [
9] reported that let-7a and miR-16 were reliable for gene expression normalization in human breast cancer studies. Let-7a, miR-17-5p, miR-103 and miR-26 were confirmed as the most stable miRNA in different specific tissues of rat [
15,
16]. In grass carp, miR-101a was showed as the most stable in all tissues and different developmental stages among the other miRNAs they studied [
17].
The Chinese perch (
Siniperca chuatsi) is one of the most commercially important carnivorous fish species in China as well as in eastern Asia [
18,
19]. In the study, the expression profile of six candidate reference miRNA genes (miR-101a, miR-146a, miR-22a, miR-23a, miR-26a and let-7a) were examined in 14 developmental stages, 8 tissues and the liver tissues from fasting-refeeding experiment of Chinese perch. U6 belongs to small nuclear RNA, and is commonly used as reference gene in miRNA quantification [
20]. A comparison assay of U6 with our candidate reference miRNAs was also conducted. The consistency of the best-scoring reference miRNA was assayed by the four statistical approaches (geNorm, BestKeeper, comparative Δ-
Ct method and NormFinder). The results revealed that all of the six miRNAs exhibited valid stability values and could be used as reference miRNAs in some of the tested conditions, and they were more suitable as reference gene for miRNA quantification than U6 in most of the tested conditions. Therefore, our study offers a comprehensive value of mRNA reference selection for RNAs expression assays at different conditions in teleost fish species.
3. Discussion
As reported in previous studies, the two most commonly used reference genes for miRNA RT-qPCR are U6 and 18S RNA [
20,
21,
22]. Because rRNAs could be expressed at much greater levels than target RNAs, bias might exist for lower-expressed target RNA expression quantification [
14,
23]. The 18S rRNA has been used as reference gene, but it also showed some variations because of its high abundance [
24]. Therefore, it is obvious that the same class of RNA should be selected for miRNA quantification. To date, study on the suitability of reference genes for miRNA expression in teleost is limited [
17].
In the study, six miRNAs (miR-22a, miR-146a, miR-101a, let-7a, miR-23a and miR-26a) were selected to assay their suitability in different tissues, different developmental stages and the fasting-refeeding treated liver tissue. The six miRNA sequences were obtained from the Chinese perch miRNA library reported by Chu [
25]. With the multiple analyses by geNorm, BestKeeper, Normfinder and Δ-
Ct, the data revealed that miR-22a, let-7a, miR-101a and miR-26a are the best reference miRNA genes based on their average expression stability (
M) or stability values. The geNorm program was used to calculate the
M stability value of a gene based on the average pairwise variation between all the studied genes. High gene expression variability results in high
M-values and exhibits low expression stability. If the
M-value of the evaluated genes is below 1.5, it indicates that the expression of the candidate gene is relatively stable [
14,
16,
17]. Our data showed that the
M-value of miR-23a, miR-146a and U6 were 1.683, 1.874 and 3.114 (
M-values > 1.5), respectively, indicating that they are not suitable as reference miRNAs in all instances. We further ran a validation experiment with comparative profiling of the miR-499 expression in white and red muscle of
Siniperca chuatsi by microarray analysis and RT-qPCR to validate our reference miRNAs. As evident from our results, inappropriate use of reference genes can significantly alter the relative expression of miR-499 between white and red muscle. When using miR-22a or let-7a as reference gene, the quantitative result was most similar to microarray analysis data between white and red muscle.
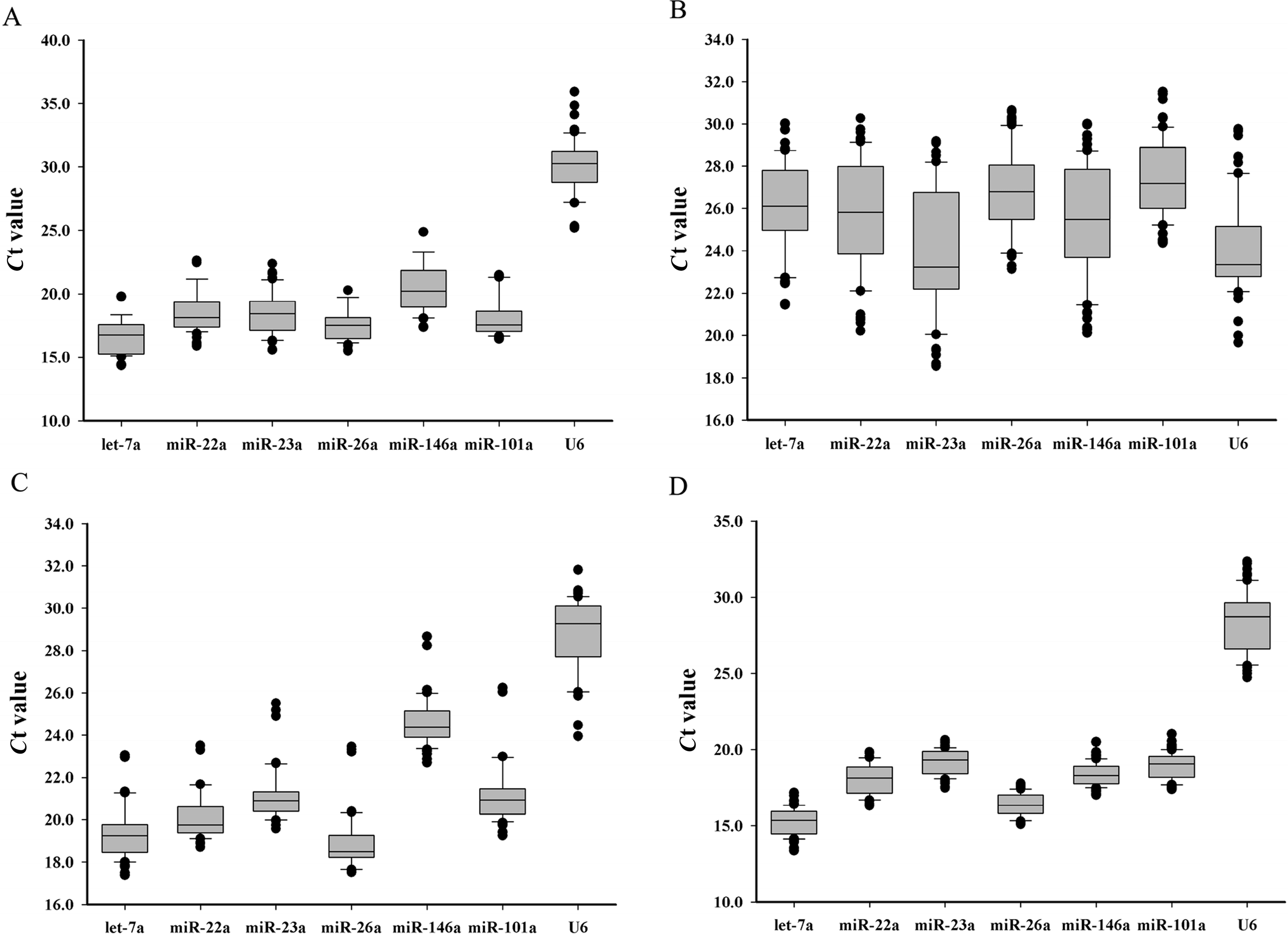
Tissue specificity may be an important factor that affects the stability of miRNA expression. In the present study, the stability of the miRNAs was evaluated for each of the eight tissues. By geNorm analysis, the stability values ranged from 0.708 to 1.447 across the different tissues (
Table 3). The results suggested that all the candidate reference miRNAs could be used for tissue-specific expression assays, but the best reference miRNA was let-7a (
Figure 2B), which was consistent to those of earlier reports. Especially, let-7a was confirmed to be a reliable reference gene in human breast and colorectal cancer [
9,
26]. Let-7a was also detected as the most stable miRNA in skeletal muscle and ovary of pig [
16,
17]. Xu [
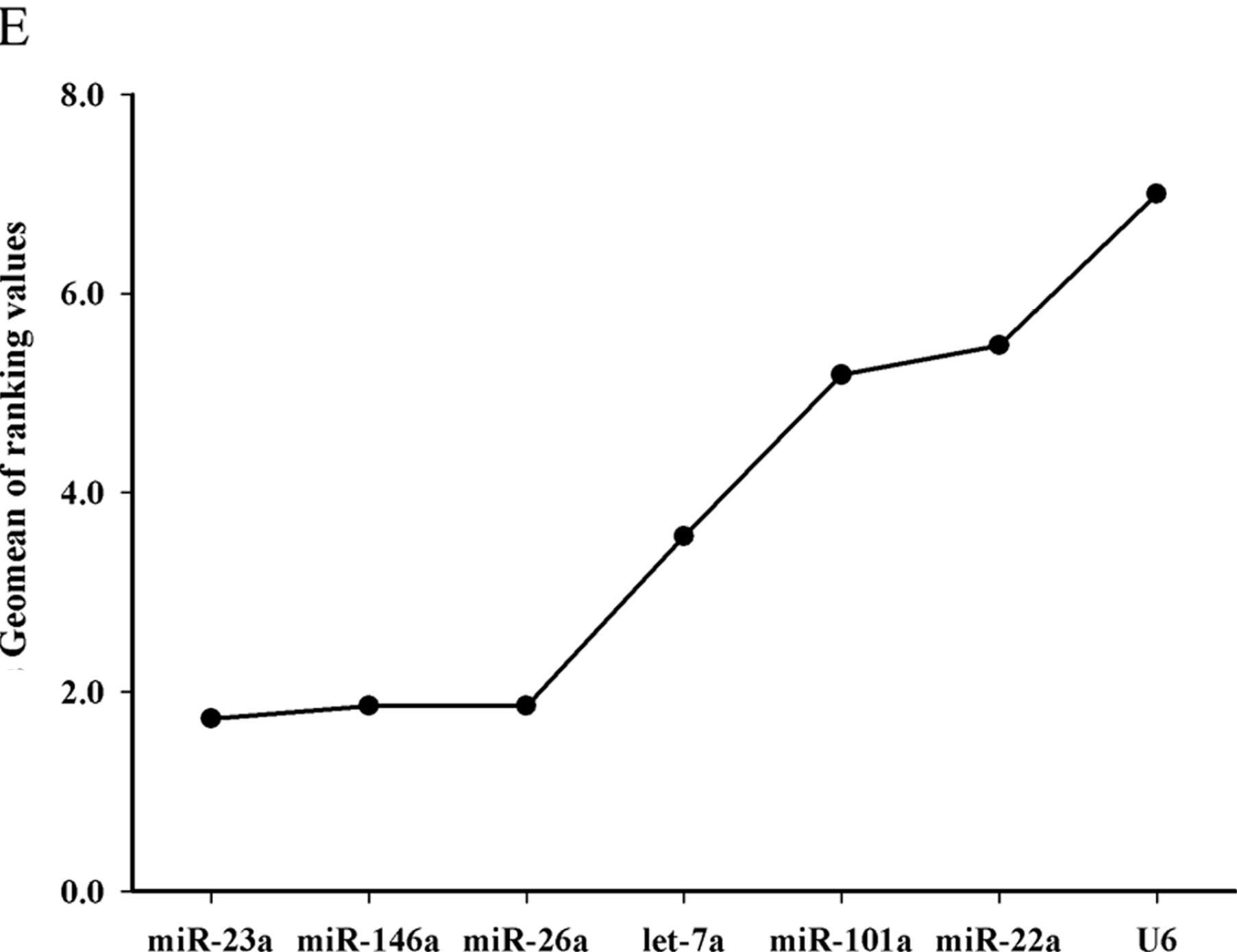
17] reported in grass carp that miR-101a was the most stable miRNA gene in the fish species among the seven miRNAs in their study. In the study, it was confirmed that let-7a, miR-26a and miR-22a were more stable and suitable than miR-101a in Chinese perch. Therefore, they could be used as suitable internal controls for miRNA expression quantification in teleost. However, the geNorm stability value of U6 was 2.23, indicating that U6 is not suitable as internal control in Chinese perch.
In the assays of the embryo samples from different developmental stages, all of the miRNAs were expressed lower than in any other samples. The cycle threshold (
Ct) values displayed a wide range from 18.54 to 31.53 (
Figure 1B). When data was compared with four algorithms (BestKeeper, geNorm, NormFinder and the comparative Δ-
Ct method), there is an apparent difference among different developmental stages, which suggested that the stability of miRNA expression appears dynamic during embryonic development. However, our results indicate that miR-101a was the most stable reference miRNA in embryonic developmental stages, followed by let-7a, miR-26a, miR-22a, miR-146a, miR-23a and U6 (
Figure 2C).
Skeletal muscle is the largest tissue in the fish body and the main edible-part for human consumption. Recent studies have shown that miRNAs play important roles in skeletal muscle development in vertebrates [
27,
28]. As earlier reported, miR-23a and let-7a were the most stable miRNA in grass carp and pig muscle at different developmental stages [
16,
17]. In the present study, the
M-values of miR-23a and let-7a were less than 1.5, indicating that they are also suitable for miRNA expression normalization in fish, at least in Chinese perch muscle.
A crucial criterion for a reference gene is that its expression levels remain constant without bias to treatment, age, tissue type, nutrition, physiological status,
etc. [
29]. Therefore, we conducted fasting-refeeding experiment to investigate the variation of six candidate reference miRNAs. All the
Ct variation ranges of these miRNAs during fasting-refeeding treatment were smaller than those in different tissues and different developmental stages. It was suggested that nutritional factors may have little effect on miRNA expression. Furthermore, based on
M-values calculated by geNorm, all of the candidate reference miRNAs were suitable for this kind of assay.
A recent study suggested that using multiple reference genes as internal controls is more accurate than using only one single gene for normalization [
14]. In the study, by analyses with Normfinder software and geNorm, the results revealed that the best combination of candidate reference genes was miR-22a and miR-23a in all conditions we assayed. The combination of miR-22a and miR-23a is the best for different tissues, miR-22a and miR-146a is the best for embryonic developmental stages, let-7a and miR-26a for post-embryonic developmental stages of muscle, and miR-26a and miR-23a for the liver tissue after nutritional treatment.
4. Materials and Methods
4.1. Fish and Tissue Sampling
All Chinese perch individuals were reared under standard conditions (22 °C, 12 h of light and dark) at the Xingda fish hatchery, Changde, Hunan, China. Fish tissue samples, including brain, kidney, liver, spleen, heart, intestine, red muscle and white muscle were collected from ten adult individuals (average body weight 500 g, 150 days of post-hatching (dph)). The fast muscle was dissected from each individual fish dorsal myotome of different post-embryonic development stages (20, 30, 50, 70, 90 and 150 (dph), ten individuals from each stage). Samples from different embryonic developmental stages including the embryos at the two-cell stage, blastula stage, gastrula stage, neurula stage, tail-bud stage, muscular effect stage, heart beating stage and larval stage (10 specimens from each stage), were obtained after artificial fertilization until hatching at the Hunan Aquaculture Institute, China. All of the tissues and embryo samples were snap-frozen in liquid nitrogen and stored at −80 °C for further processing.
To investigate expression stability of the miRNAs during fasting and refeeding experiments, 80 Chinese perch juveniles (average body weight 150 g, 90 dph) in a net cage (5 m × 5 m × 2 m) were reared under the same conditions as described above and they were fed with mud carp (average body weight 10 g) for one week. The fish juveniles were fasted for one week, and then fed with a single meal (mud carp). Liver samples were collected at 0 h (before the recovery meal), 1, 3, 6, 12, 24, 48 and 96 h after the single meal feeding respectively (ten individuals from each stage).
4.2. RNA Isolation and First Strand cDNA Synthesis
All tissue samples were ground in liquid nitrogen as preprocesses. The total RNAs were extracted from embryos and tissue samples using TRIzol® Reagent (Invitrogen, Waltham, MA, USA) according to the manufacturer’s protocol, and then treated with RNAse-free DNAse I (Promega, Fitchburg, WI, USA) in the presence of RNAse inhibitor (Sigma, Saint Louis, MO, USA). The RNAs were harvested by ethanol precipitation and their concentration and purity were determined by at the absorbance at 260 and 280 nm in a Nanodrop 2000 spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA). The RNA integrity was assessed by the 18S and 28S band intensity ratio after 1.5% agarose gel electrophoresis visualized by GoldView II staining (Beijing, China). Equal amounts of 800 ng/μL of RNAs were polyadenylated by poly (A) polymerase, and then reverse transcribed using the One Step PrimeScript miRNA cDNA synthesis kit (TaKaRa, Dalian, China) with the Universal Adaptor Primer (a poly (T) primer ligated with an adapter) for miRNA quantitative assays.
4.3. Quantitative Real-Time PCR Assay
The forward primers for miRNAs RT-qPCR were designed based on the mature miRNA sequence (
Table 1), while about 2 to 6 bp was removed at the 3' end and about 2 to 6 bp was added at 5' end according to the Tm value. The primers for U6 RT-qPCR referred to the previous report [
30]. Standard curves were generated to calculate the RT-qPCR efficiency. All standard curves were made using 10 fold serial dilutions from a pool of cDNA which consist of cDNA samples from eight tissues (including brain, kidney, liver, spleen, heart, intestine, red muscle and white muscle,
n = 80). The cDNA samples were used as templates for quantitative RT-PCR assays with SYBR Premix Ex Taq™ II (TaKaRa, Dalian, China) and its amplification reaction was carried out with Bio-Rad CFX96 system (Bio-Rad, Hercules, CA, USA). Each of 2 μL cDNA template was added to a total volume of 25 μL reaction mix containing 12.5 μL SYBR Green mix, 1 μL of miRNA specific forward primer and 1 μL of miRNA universal downstream primer forward primer (
Table 1, 10 μmol/L), 8.5 μL nuclease-free water. The following protocol was used: (i) Pre-denaturation at 95 °C for 60 s; (ii) Amplification and quantification, 40 repeated cycles at 95 °C for 5 s and at 60 °C for 25 s; (iii) Melting curve program (65–95 °C with heating rate of 0.1 °C/s and fluorescence measurement).
4.4. Selection of Candidate Reference miRNAs and Expression Stability Analyses
The candidate reference miRNAs selected for the evaluation include: miR-101a, miR-146a, miR-22a, miR-23a, miR-26a and let-7a; These candidate miRNAs were previously evaluated as reference miRNAs [
16,
17,
31]. The stability of each candidate miRNA was analyzed with geNorm [
14], NormFinder [
32], BestKeeper [
33] and the comparative Δ-
Ct [
34] method algorithm. The geNorm algorithm was used to calculate the gene expression stability (
M) value based on average pairwise variation between all studied genes. NormFinder was used to analysis of variance (ANOVA) mathematical model and estimates of intra- and inter-group variation. In both programs, the lowest stability values indicate the most stably expressed reference genes, which allow them to be ranked according to expression stability. Based on the rankings from each program, RefFinder (
http://www.leonxie.com/referencegene.php) assigns an appropriate value to an individual gene with overall final ranking. Then we ran a validation experiment with comparative profiling of the miR-499 expression in white muscle and red muscle of
Siniperca chuatsi by microarray analysis and RT-qPCR; For the method of miRNA microarray analysis, please refer to Chu
et al. [
25].