Characterization and Structure Prediction of Partial Length Protein Sequences of pcoA, pcoR and chrB Genes from Heavy Metal Resistant Bacteria from the Klip River, South Africa
Abstract
:1. Introduction
2. Results and Discussion
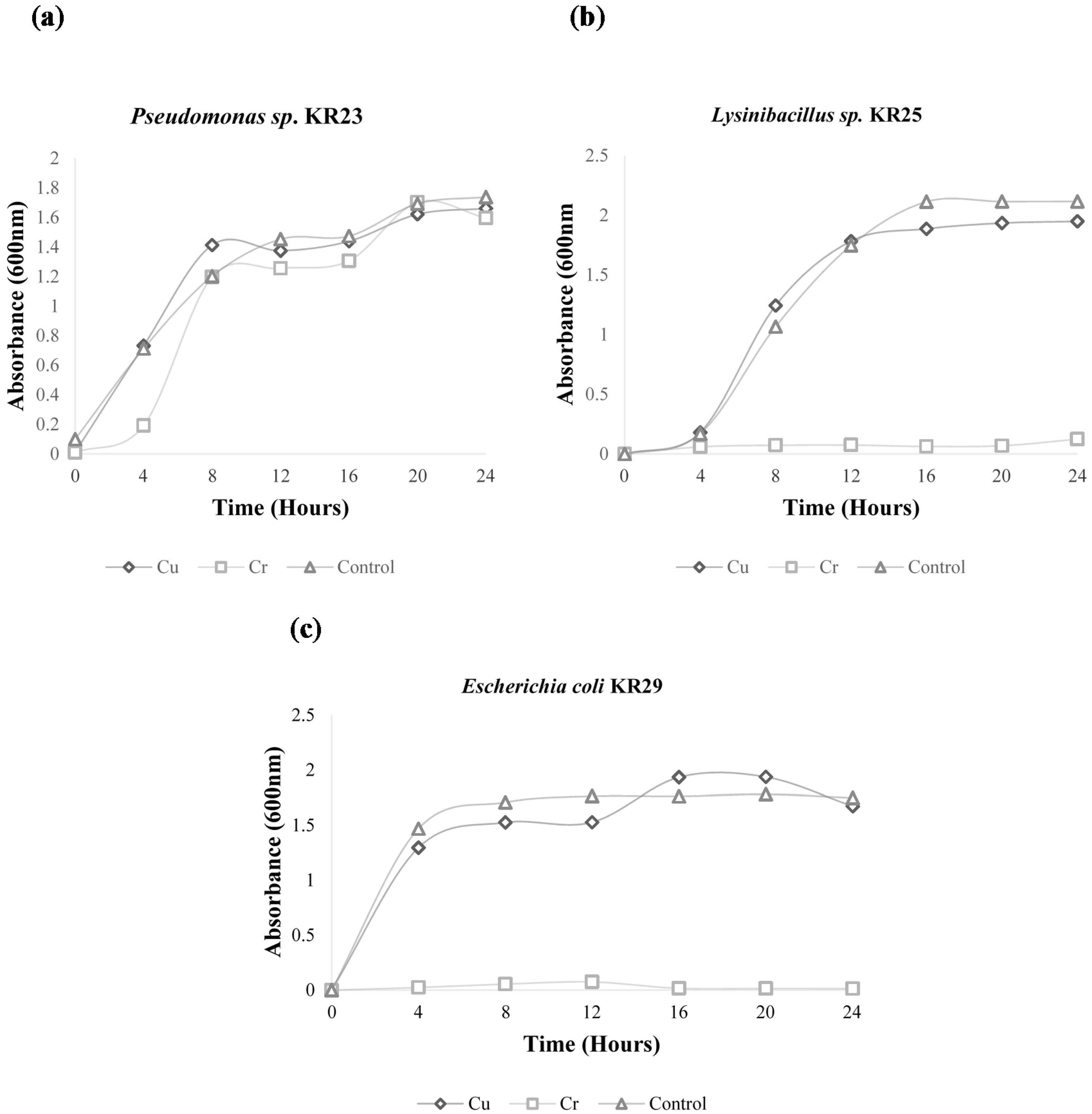
2.1. Growth Curve Analysis
2.2. Amplification of Heavy Metal Resistance Genes

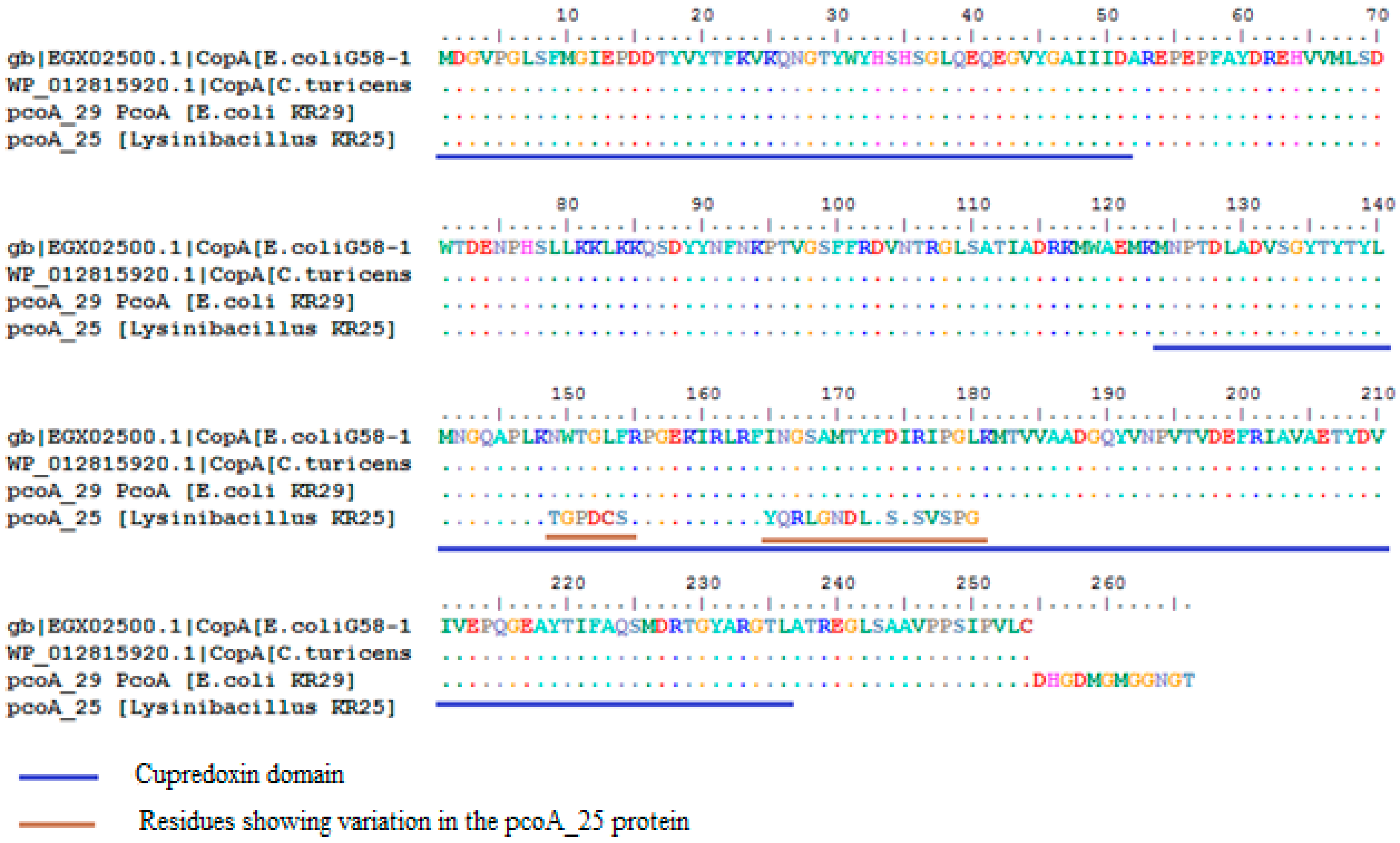
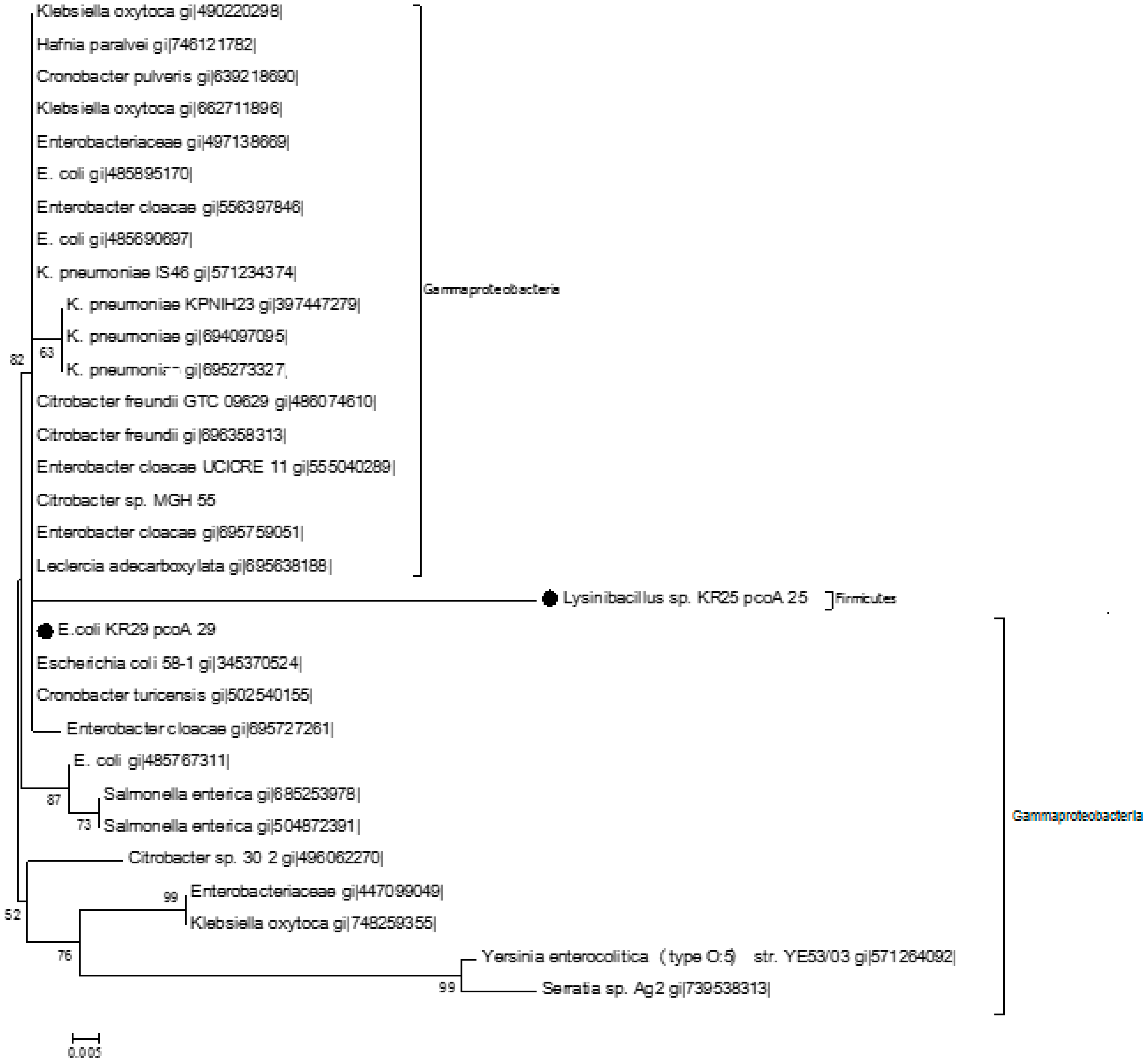
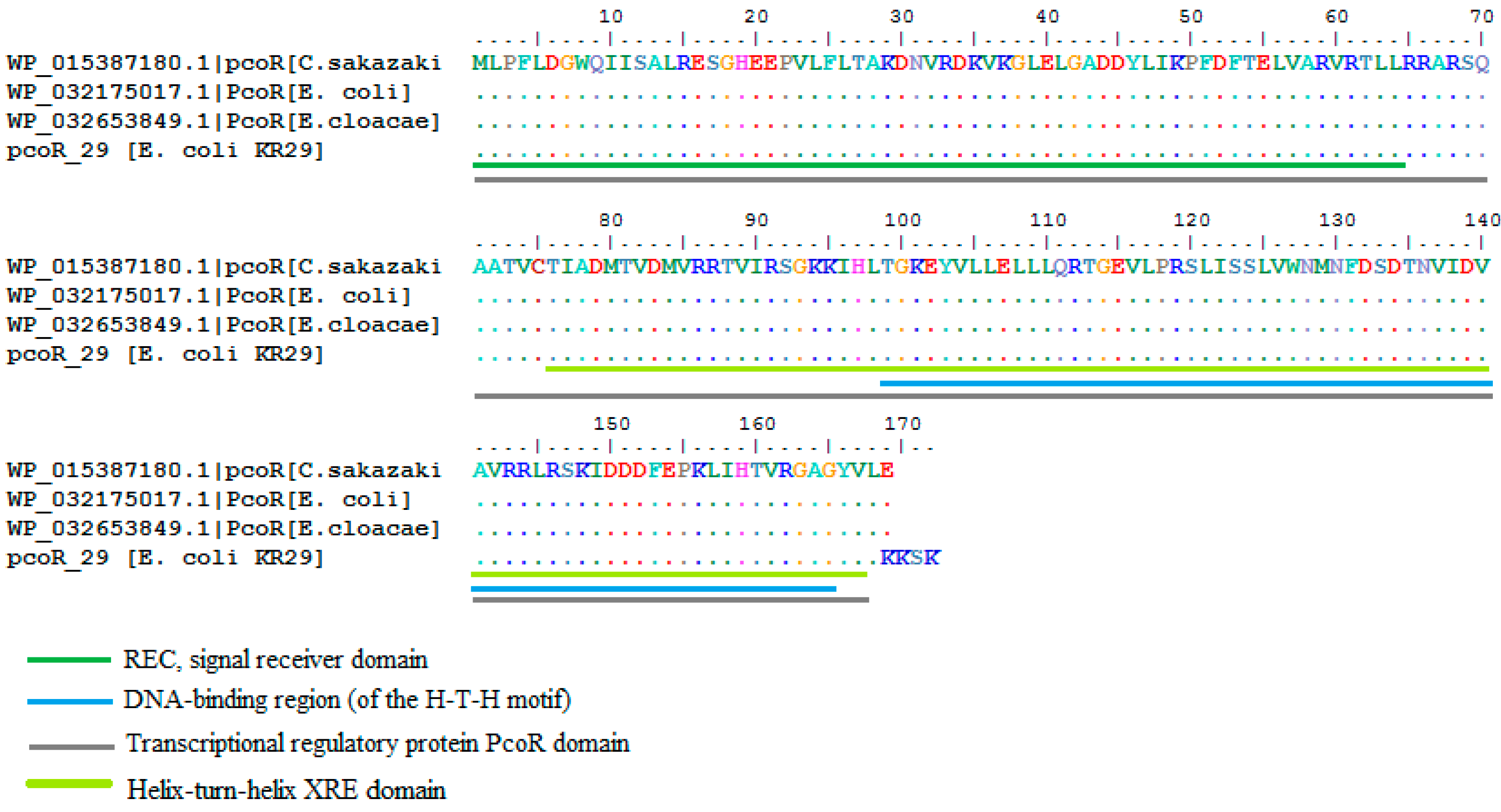
2.3. Homology Analysis of Amplified Genes


| Properties | ChrB_23 | pcoA_25 | pcoA_29 | pcoR_29 |
|---|---|---|---|---|
| Number of amino acids | 95 | 180 | 266 | 172 |
| Molecular weight (units) | 10,558.9 | 20,544.1 | 29,571.6 | 19,565.8 |
| Theoretical Pi | 4.75 | 5.80 | 5.16 | 9.44 |
| Total number of negatively charged residues (Asp + Glu) | 12 | 23 | 33 | 23 |
| Total number of negatively charged residues (Arg + Lys) | 8 | 20 | 25 | 28 |
| Extinction coefficient | 12,950 | 34,380 | 45,840 | 15,470 |
| Extinction coefficient * | - | 34,380 | 45,840 | 15,470 |
| Instability Index | 12.158 | 25.06 | 20.63 | 41.56 |
| Aliphatic index | 74 | 63.89 | 70.38 | 109.30 |
| Grand average of hydropathicity | −0.323 | −0.612 | −0.364 | −0.103 |
2.4. I-TASSER Structural and Functional Prediction Results
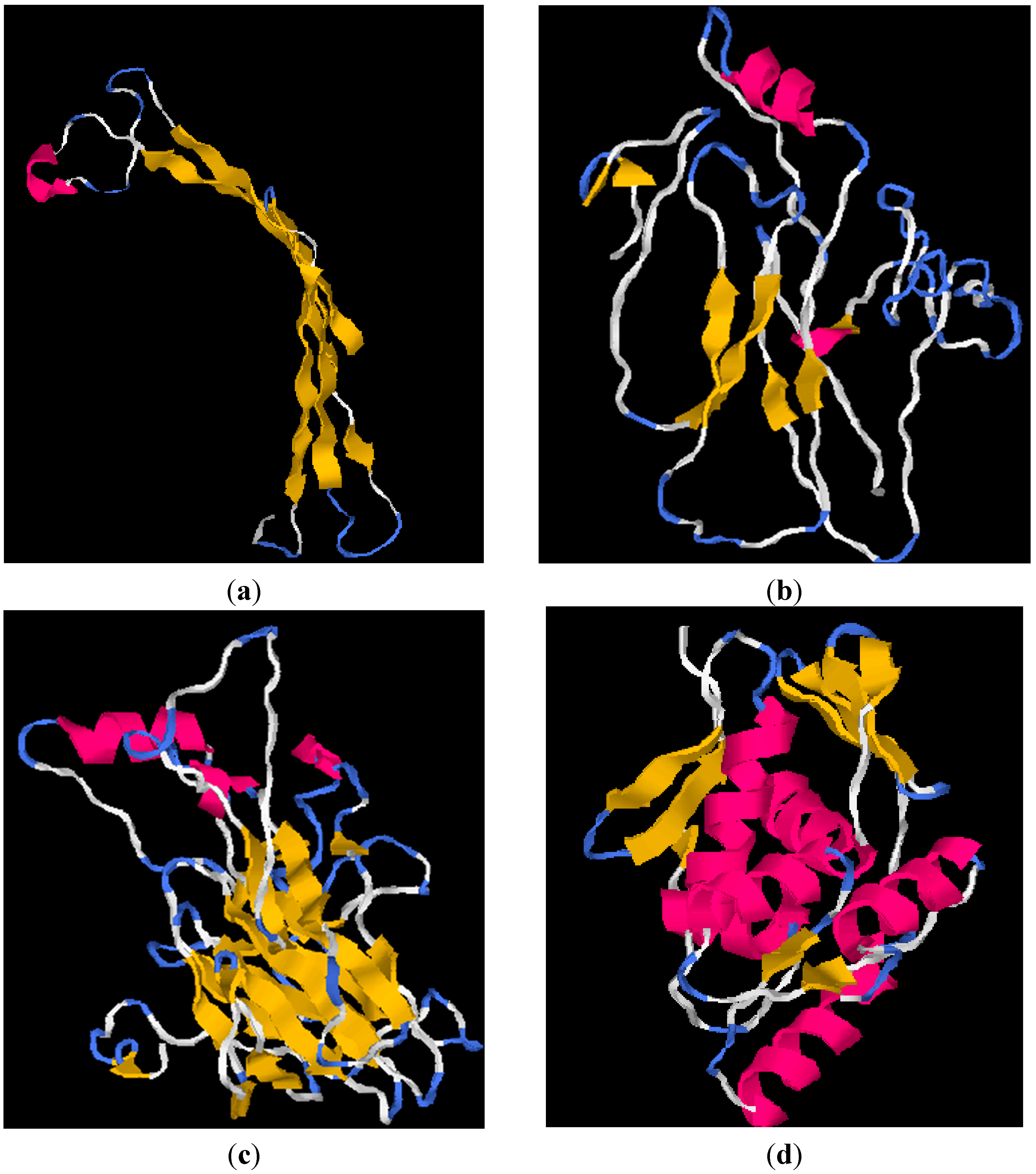
| Properties | chrB_23 | pcoA_25 | pcoA_29 | pcoR_29 |
|---|---|---|---|---|
| C-score | −1.34 | −0.94 | −0.02 | 1.20 |
| TM-score | 0.55 ± 0.15 | 0.60 ± 0.14 | 0.71 ± 0.12 | 0.88 ± 0.07 |
| Root Mean Square Deviation (Å) | 6.6 ± 4.0 | 7.1 ± 4.2 | 6.0 ± 3.7 | 2.7 ± 2.0 |
2.5. Ramachandran Plot Analysis
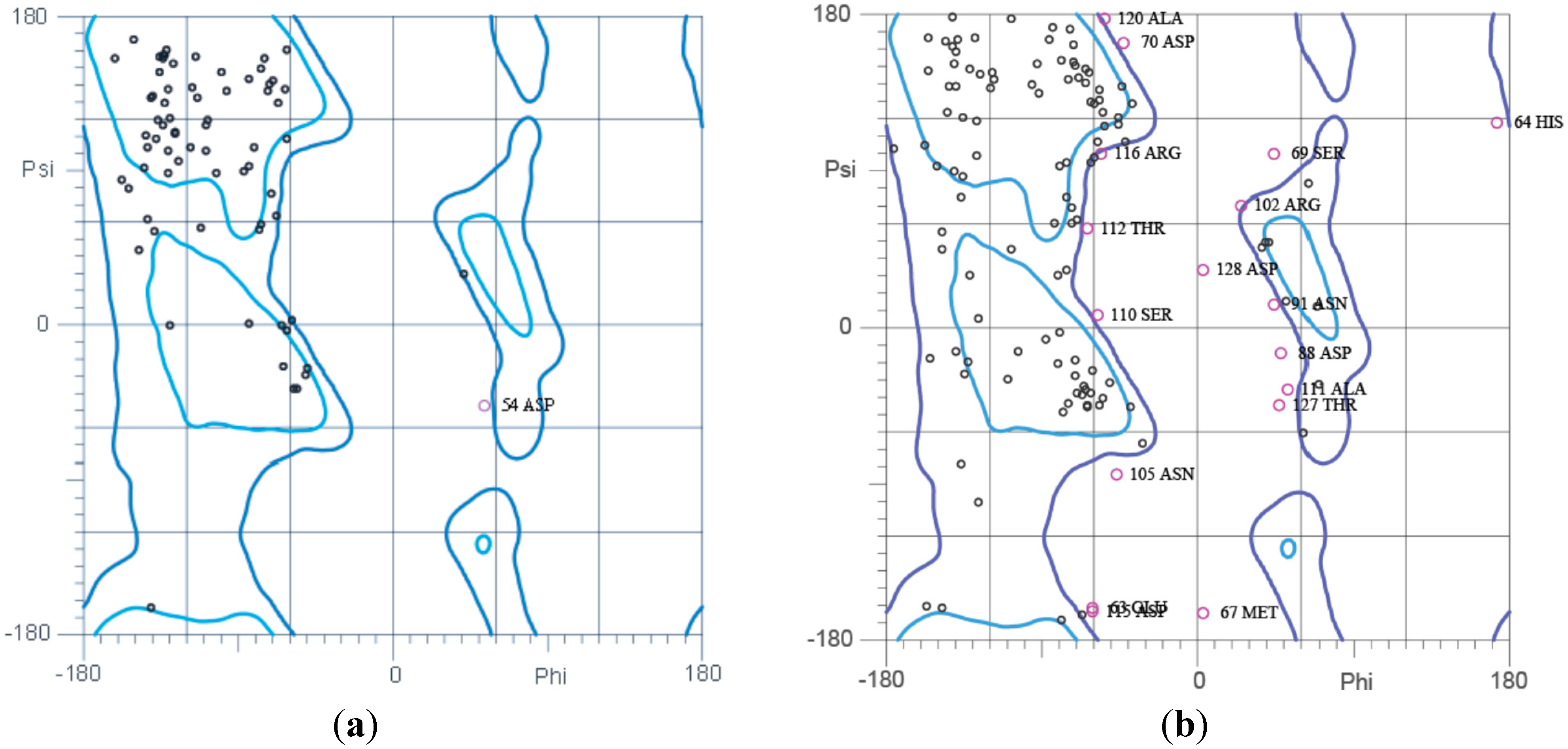
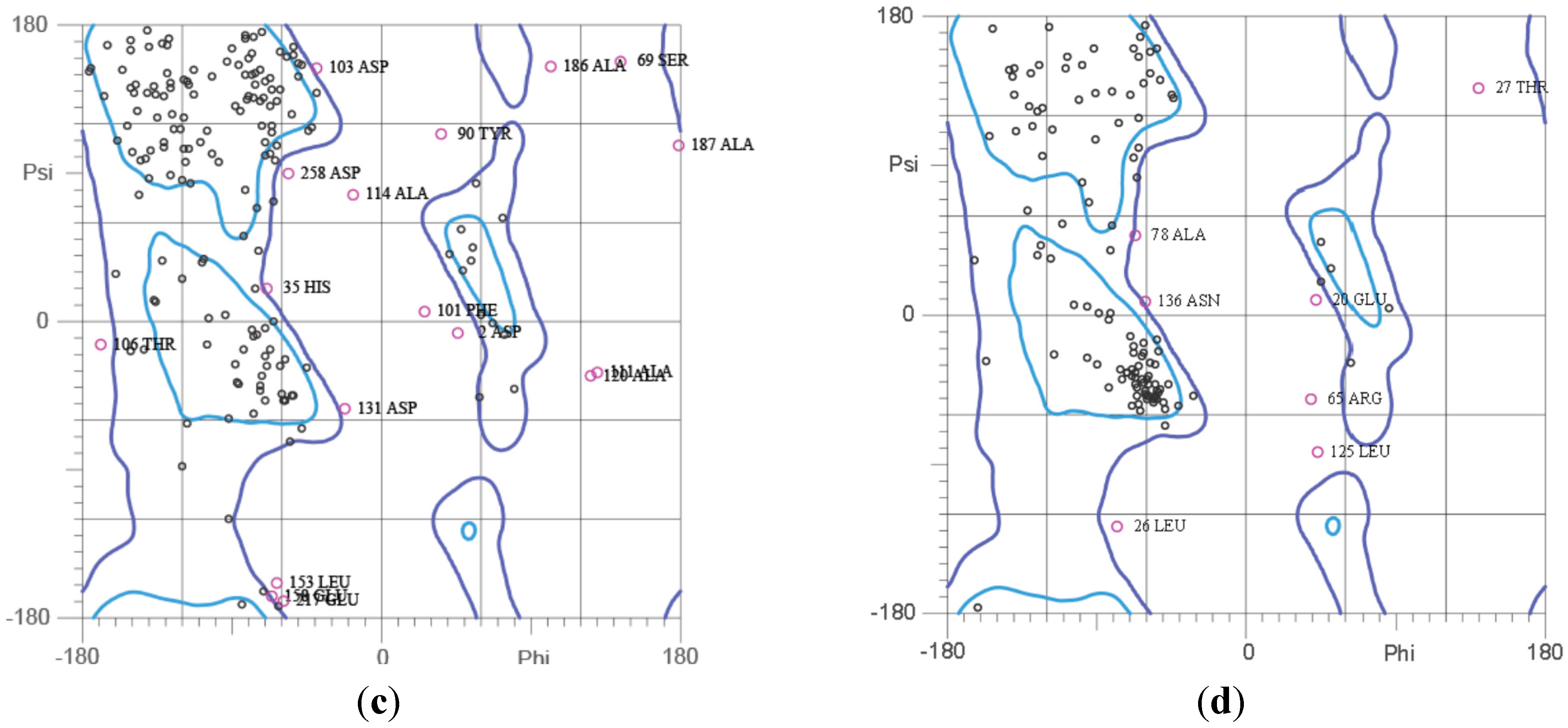
2.6. Plasmid Profiling and Curing

| Antibiotic | KR23 | KR25 | KR29 |
|---|---|---|---|
| Streptomycin | NZ | NZ | 16(S) |
| Cephalothin acid | NZ | 27(S) | 13(S) |
| Tobramycin | 20(S) | 18(S) | 18(S) |
| Neomycin | 20(S) | 17(S) | 13(S) |
| Tetracycline | 23(S) | 21(S) | 19(S) |
| Cotrimoxazole | 28(S) | 22(S) | 22(S) |
| Vancomycin | NZ | 18(S) | NZ |
| Ampicillin | NZ | 27(S) | 14(S) |
| Amoxicillin | NZ | 30(S) | 12(R) |
| Antibiotic | KR23 | KR25 | KR29 |
|---|---|---|---|
| Streptomycin | NZ | NG | 18(S) |
| Cephalothin acid | NZ | NG | 16(S) |
| Tobramycin | 31(S) | NG | 16(S) |
| Neomycin | 15(S) | NG | 15(S) |
| Tetracycline | 30(S) | NG | 23(S) |
| Cotrimoxazole | 33(S) | NG | 24(S) |
| Vancomycin | NZ | NG | NZ |
| Ampicillin | NZ | NG | 17(S) |
| Amoxicillin | NZ | 3G | 15(S) |
| Heavy metal resistance profiles after curing | |||
| Control | +++ | +++ | +++ |
| Chromate (0.2mm) | ++ | NG | NG |
| Copper (0.6mm) | ++ | NG | ++ |
3. Discussion
3.1. Chromate Resistance Genes
3.2. Copper Resistance Genes
3.3. Plasmid Isolation and Curing
3.4. Physico-Chemical Properties
3.5. Protein Structures
4. Experimental Section
4.1. Growth Studies
4.2. Genomic DNA Extraction
| Strain | Source | Location | Accession Number |
|---|---|---|---|
| Pseudomonas KR23 | Klip River (water sample) | South Africa | KJ935916 |
| Lysinibacillus KR25 | Klip River (water sample) | South Africa | KJ935917 |
| Escherichia coli | Klip River (water sample) | South Africa | KJ935918 |
4.3. Amplification of Heavy Metal Resistant Genes
| Gene to Be Amplified | Primer Sequences | Orientation | Corresponding Microbe with Heavy Metal Resistance Gene | Exact Length of Amplified Region |
|---|---|---|---|---|
| chrB | GTCGTTAGCTTGCCAACATC | Forward | C. metallidurans CH34 | 450 [14] |
| CGGAAAGCAAGATGTCGATCG | Reverse | |||
| pcoA | CGTCTCGACGAACTTTCCTG | Forward | E. coli E8739 | 1791 [6] |
| GGACTTCACGAAACATTCCC | Reverse | Plasmid pPA87 | ||
| pcoR | CAGGTCGTTACCTGCAGCAG | Forward | E. coli E8739 | 636 [6] |
| CTCTGATCTCCAGGACATATC | Reverse | Plasmid pPA87 |
4.4. Gel Electrophoresis
4.5. PCR Product Purification and Sequencing of pcoA, pcoR and chrB Genes
4.6. Homology Analysis of Amplified Genes
4.7. Isolation of Plasmid DNA
4.8. Plasmid Curing
4.9. Antibiotic Profiles of Wild Type Strains and Cured Derivatives
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ozer, G.; Ergene, A.; Icgen, B. Biochemical and molecular characterization of strontium-resistant environmental isolates of Pseudomonas fluorescens and Sphingomonas paucimobilis. Geomicrobiol. J. 2013, 30, 381–390. [Google Scholar] [CrossRef]
- Mccarthy, T.S.; Arnold, V.; Venter, J.; Ellery, W.N. The collapse of the Johannesburg Klip River Wetland. S. Afr. J. Sci. 2007, 103, 391–397. [Google Scholar]
- Chihomvu, P.; Stegmann, P.; Pillay, M. Identification and Characterization of Heavy Metal Resistant Bacteria from the Klip River. In Proceedings of the International Conference on Ecological, Environmental and Biological Sciences, WASET, Cape Town, South African, 25–26 November 2014.
- Aktan, Y.; Tan, S.; Icgen, B. Characterization of lead-resistant river isolate Enterococcus faecalis and assessment of its multiple metal and antibiotic resistance. Environ. Monit. Assess. 2013, 185, 5285–5293. [Google Scholar] [CrossRef] [PubMed]
- Macomber, L.; Imlay, J. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc. Natl. Acad. Sci. USA 2009, 106, 8344–8349. [Google Scholar] [CrossRef] [PubMed]
- Brown, N.L.; Rouch, D.A.; Lee, B.T.O. Copper resistance systems in bacteria. Plasmid 1992, 27, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Silver, S.; Lee, B.T.O.; Brown, N.L.; Cooksey, D.A. Bacterial plasmid resistances to copper, cadmium and zinc. In Chemistry of Copper and Zinc Triads; Welch, A.J., Ed.; The Royal Society of Chemistry: London, UK, 1993; pp. 33–53. [Google Scholar]
- Cooksey, D.A. Genetics of bactericide resistance in plant pathogenic bacteria. Annu. Rev. Phytopathol. 1990, 28, 201–219. [Google Scholar] [CrossRef]
- Huffman, D.; Huyett, J.; Outten, F.; Doan, P.; Finney, L.; Hoffman, B.; O’Halloran, T.V. Spectroscopy of Cu (II)-PcoC and the multicopper oxidase function of PcoA, two essential components of Escherichia coli pco copper resistance operon. Biochemistry 2002, 41, 10046–10055. [Google Scholar] [CrossRef] [PubMed]
- Djoko, K.; Chong, L.; Wedd, A.; Xiao, Z. Reaction mechanisms of the multicopper oxidase CueO from Escherichia coli support its functional role as a cuprous oxidase. J. Am. Chem. Soc. 2010, 132, 2005–2015. [Google Scholar] [CrossRef] [PubMed]
- Silver, S. Bacterial resistances to toxic metal ions—A review. Gene 1996, 179, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Badar, U.; Ahmed, N.; Shoeb, E.; Gadd, G. Identification of the pco operon in Enterobacter species isolated from contaminated soil. Int. J. 2014, 2, 227–233. [Google Scholar]
- Ramirez-Diaz, M.; Diaz-Perez, C.; Vargas, E.; Riveros-Rosas, H.; Campos-Garcia, J.; Cervantes, C. Mechanisms of bacterial resistance to chromium compounds. Biometals 2008, 21, 321–332. [Google Scholar] [CrossRef] [PubMed]
- Nies, A.; Nies, D.H.; Silver, S. Nucleotide sequence and expression of a plasmid encoded chromate resistance determinant from Alcaligens eutrophus. J. Biol. Chem. 1990, 265, 5648–5653. [Google Scholar] [PubMed]
- Aguilar-Barajas, E.; Paluscio, E.; Cervantes, C.; Rensing, C. Expression of chromate resistance genes from Shewanella sp. strain ANA-3 in Escherichia coli. FEMS Microbiol. Lett. 2008, 285, 97–100. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Perez, C.; Cervantes, C.; Campos-Garcia, J.; Julian-Sanchez, A.; Riveros-Rosas, H. Phylogenetic analysis of the chromate ion transporter (CHR) superfamily. FEBS J. 2007, 274, 6215–6227. [Google Scholar] [CrossRef] [PubMed]
- Branco, R.; Morais, P. Identification and characterization of the transcriptional regulator ChrB in the chromate resistance determinant of Ochrobactrum tritici 5bvl1. PLoS ONE 2013, 8, e77987. [Google Scholar] [CrossRef] [PubMed]
- Bopp, L.; Chakrabarty, A.; Ehrlich, H. Chromate resistance plasmid in Pseudomonas fluorescens. J. Bacteriol. 1983, 155, 1105–1109. [Google Scholar] [PubMed]
- Efstathiou, J.; McKay, L. Inorganic salts resistance associated with a lactose-fermenting plasmid in Streptococcus lactis. J. Bacteriol. 1977, 130, 257–265. [Google Scholar] [PubMed]
- Henne, K.; Nakatsu, C.; Thompson, D.; Konopka, A. High-level chromate resistance in Arthrobacter sp. strain FB24 requires previously uncharacterized accessory genes. BMC Microbiol. 2009, 9, 199. [Google Scholar] [CrossRef] [PubMed]
- Kamika, I.; Momba, M. Assessing the resistance and bioremediation ability of selected bacterial and protozoan species to heavy metals in metal-rich industrial wastewater. BMC Microbiol. 2013, 13, 28. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y. I-TASSER server for protein 3D structure prediction. BMC Bioinform. 2008, 9, 40. [Google Scholar] [CrossRef]
- Laurenzi, A.; Hung, L.; Samudrala, R. Structure prediction of partial-length protein sequences. Int. J. Mol. Sci. 2013, 14, 14892–14907. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.; Thornton, J. Searching for functional sites in protein structures. Curr. Opin. Chem. Biol. 2004, 8, 3–7. [Google Scholar] [CrossRef] [PubMed]
- Watson, J.; Sanderson, S.; Ezersky, A.; Savchenko, A.; Edwards, A.; Orengo, C.; Joachimiak, A.; Laskowski, R.A.; Thornton, J.M. Towards fully automated structure-based function prediction in structural genomics: A case study. J. Mol. Biol. 2007, 367, 1511–1522. [Google Scholar] [CrossRef] [PubMed]
- Bouanchaud, D.; Scavizzi, M.; Chabbert, Y. Elimination by ethidium bromide of antibiotic resistance in enterobacteria and staphylococci. J. Gen. Microbiol. 1968, 54, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Spengler, G.; Molnar, A.; Schelz, Z.; Amaral, L.; Sharples, D.; Molnar, J. The mechanism of plasmid curing in bacteria. Curr. Drug Targets 2006, 7, 823–841. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Lu, S.; Anderson, J.; Chitsaz, F.; Derbyshire, M.; DeWeese-Scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzalez, N.R.; et al. CDD: A Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011, 39, 225–229. [Google Scholar] [CrossRef] [PubMed]
- Altenhoff, A.M.; Škunca1, N.; Glover, N.; Train, C.; Sueki, A.; Piližota1, I.; Gori, K.; Tomiczek, B.; Müller, S.; Redestig, H.; et al. The OMA orthology database in 2015: Function predictions, better plant support, synteny view and other improvements. Nucleic Acids Res. 2014. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: New York, NY, USA, 2000. [Google Scholar]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Wilkins, M.; Appel, R.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server; Humana Press: Clifton, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Davis, I.W.; Leaver-Fay, A.; Chen, V.B.; Block, J.N.; Kapra, G.J.; Wang, X.; Murray, L.W.; Arendall, W.B., III; Snoeyink, J.; Richardson, J.S.; et al. MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007, 35, W375–W383. [Google Scholar] [CrossRef] [PubMed]
- Raja, E.C.; Selvam, G.S.; Omnie, K. Isolation, identification and characterization of heavy metal resistant bacteria from sewage. Int. Jt. Symp. Geodisaster Prev. Geoenviron. Asia 2009, 205–211. [Google Scholar]
- Fan, B.; Grass, G.; Rensing, C.; Rosen, B.P. Escherichia coli CopA N-terminal Cys(X)(2)Cys motifs are not required for copper resistance or transport. Biochem. Biophys. Res. Commun. 2001, 286, 414–418. [Google Scholar]
- Tetaz, T.; Luke, R. Plasmid-controlled resistance to copper in Escherichia coli. J. Bacteriol. 1983, 154, 1263–1268. [Google Scholar] [PubMed]
- Mellano, M.; Cooksey, D. Induction of the copper resistance operon from Pseudomonas syringae. J. Bacteriol. 1988, 170, 4399–4401. [Google Scholar] [PubMed]
- Monchy, S.; Benotmane, M.; Janssen, P.; Vallaeys, T.; Taghavi, S.; van der Lelie, D.; Mergeay, M. Plasmids pMOL28 and pMOL30 of Cupriavidus metallidurans are specialized in the maximal viable response to heavy metals. J. Bacteriol. 2007, 189, 7417–7425. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Ma, Z.; Liang, J.; Li, H.; Wang, E.; Wei, G. Characterization of a copper-resistant symbiotic bacterium isolated from Medicago lupulina growing in mine tailings. Bioresour. Technol. 2011, 102, 703–709. [Google Scholar] [CrossRef] [PubMed]
- Brown, N.; Barrett, S.; Camakaris, J.; Lee, B.; Rouch, D. Molecular genetics and transport analysis of the copper-resistance determinant (pco) from Escherichia coli plasmid pRJ1004. Mol. Microbiol. 1995, 17, 1153–1166. [Google Scholar] [CrossRef] [PubMed]
- Rouch, D.; Brown, N. Copper-inducible transcriptional regulation at two promoters in the Escherichia coli copper resistance determinant pco. Microbiology 1997, 143, 1191–1202. [Google Scholar] [CrossRef] [PubMed]
- Altimira, F.; Yáñez, C.; Bravo, G.; González, M.; Rojas, L.; Seeger, M. Characterization of copper-resistant bacteria and bacterial communities from copper-polluted agricultural soils of central Chile. BMC Microbiol. 2012, 12, 193. [Google Scholar] [CrossRef] [PubMed]
- Frost, L.; Leplae, R.; Summers, A.; Toussaint, A. Mobile genetic elements: The agents of open source evolution. Nat. Rev. Microbiol. 2005, 3, 722–732. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, S.; Chaslus-Dancla, E. Use of antimicrobials in veterinary medicine and mechanisms of resistance. Vet. Res. 2001, 32, 201–225. [Google Scholar] [CrossRef] [PubMed]
- Fraser, C.; Eisen, J.; Salzberg, S. Microbial genome sequencing. Nature 2000, 406, 799–803. [Google Scholar] [CrossRef] [PubMed]
- Clermont, D.; Horaud, T. Identification of chromosomal antibiotic resistance genes in Streptococcus anginosus (“S. milleri”). Antimicrob. Agents Chemother. 1990, 34, 1685–1690. [Google Scholar] [CrossRef] [PubMed]
- Jatav, V.; Parasher, L.; Raje, A.; Tiwari, S. Homology Modeling and Analysis of Structure Predictions of KLF8 Protein From Homo Sapiens. Int. J. Pharm. Sci. Res. 2014, 5, 1045–1050. [Google Scholar]
- Kyte, J.; Doolittle, R. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Bruns, M.A.; Tiedje, J.M. DNA recovery from Soils of Diverse Composition. Appl. Environ. Microbiol. 1996, 62, 316–322. [Google Scholar] [PubMed]
- Sanger, F.; Coulson, A. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J. Mol. Biol. 1975, 94, 441–448. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.; Madden, T.; Sch’affer, A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Birnboim, H.; Doly, J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979, 7, 1513–1523. [Google Scholar] [CrossRef] [PubMed]
- Malik, A.; Jaiswal, R. Metal resistance in Pseudomonas strains isolated from soil treated with industrial wastewater. World J. Microbiol. Biotechnol. 2000, 16, 177–182. [Google Scholar] [CrossRef]
- Ginns, C.; Benham, M.; Adams, L.; Whithear, K.; Bettelheim, K.; Crabb, B.; Browning, G.F. Colonization of the respiratory tract by a virulent strain of avian Escherichia coli requires carriage of a conjugative plasmid. Infect. Immun. 2000, 68, 1535–1541. [Google Scholar] [CrossRef] [PubMed]
- Bauer, W.; Kirby, W.M.; Sherris, J.C.; Turck, M. Antibiotic Susceptibility Testing by a standardized single disc method. Am. J. Clin. Pathol. 1966, 36, 493–496. [Google Scholar]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chihomvu, P.; Stegmann, P.; Pillay, M. Characterization and Structure Prediction of Partial Length Protein Sequences of pcoA, pcoR and chrB Genes from Heavy Metal Resistant Bacteria from the Klip River, South Africa. Int. J. Mol. Sci. 2015, 16, 7352-7374. https://doi.org/10.3390/ijms16047352
Chihomvu P, Stegmann P, Pillay M. Characterization and Structure Prediction of Partial Length Protein Sequences of pcoA, pcoR and chrB Genes from Heavy Metal Resistant Bacteria from the Klip River, South Africa. International Journal of Molecular Sciences. 2015; 16(4):7352-7374. https://doi.org/10.3390/ijms16047352
Chicago/Turabian StyleChihomvu, Patience, Peter Stegmann, and Michael Pillay. 2015. "Characterization and Structure Prediction of Partial Length Protein Sequences of pcoA, pcoR and chrB Genes from Heavy Metal Resistant Bacteria from the Klip River, South Africa" International Journal of Molecular Sciences 16, no. 4: 7352-7374. https://doi.org/10.3390/ijms16047352





