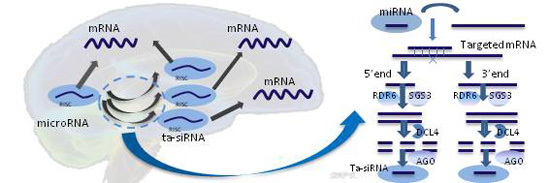
Predicted Trans-Acting siRNAs in the Human Brain
Abstract
:1. Introduction
2. Results and Discussion
2.1. A. thaliana ta-siRNA Prediction Analysis
| Predicted TAS Genes a | ta-siRNA Sequence | TAS Gene Description b | References |
|---|---|---|---|
| AT1G63400 | TGATCAAAAGGTCTATA | pentatricopeptide repeat protein (PPR) | Prediction [24,26] |
| AT1G62930 | TGATCAAAAGGTCTATA | RNA processing factor 3, RPF3 | Prediction [24,30] |
| AT1G63130 | TGATCAAAAGGTCTATA | Tetratricopeptide repeat (TPR)-like superfamily protein | TAS2 [9,11,12,13,24,26,27,29,31] |
| AT1G62910 | TGATCAAAAGGTCTATA | pentatricopeptide repeat protein (PPR) | Prediction [24,26] |
| AT1G63150 | TGTGGAAGTTGCTGTTG | pentatricopeptide repeat protein (PPR) | TAS2 [13,24,26,28,29] |
| TGATCTTCAACACAATC | pentatricopeptide repeat protein (PPR) | TAS2 [13,24,26,28,29] | |
| AT2G39675 | GTTGCTAATACAGTTAC | TAS1C; other RNA | TAS1C [11,12,24,26,32,33] |
| TCTAAGTCCAACATAGC | TAS1C; other RNA | TAS1C [11,12,24,26,32,33] | |
| TTCTAAGTCCAACATAG | TAS1C; other RNA | TAS1C [11,12,24,26,32,33] | |
| AT2G27400 | TTCTAAGTCCAACATAG | TAS1A; other RNA | TAS1A [11,19,26,33,34,35,36,37] |
| TCTAAGTCCAACATAGC | TAS1A; other RNA | TAS1A [11,19,26,33,34,35,36,37] |
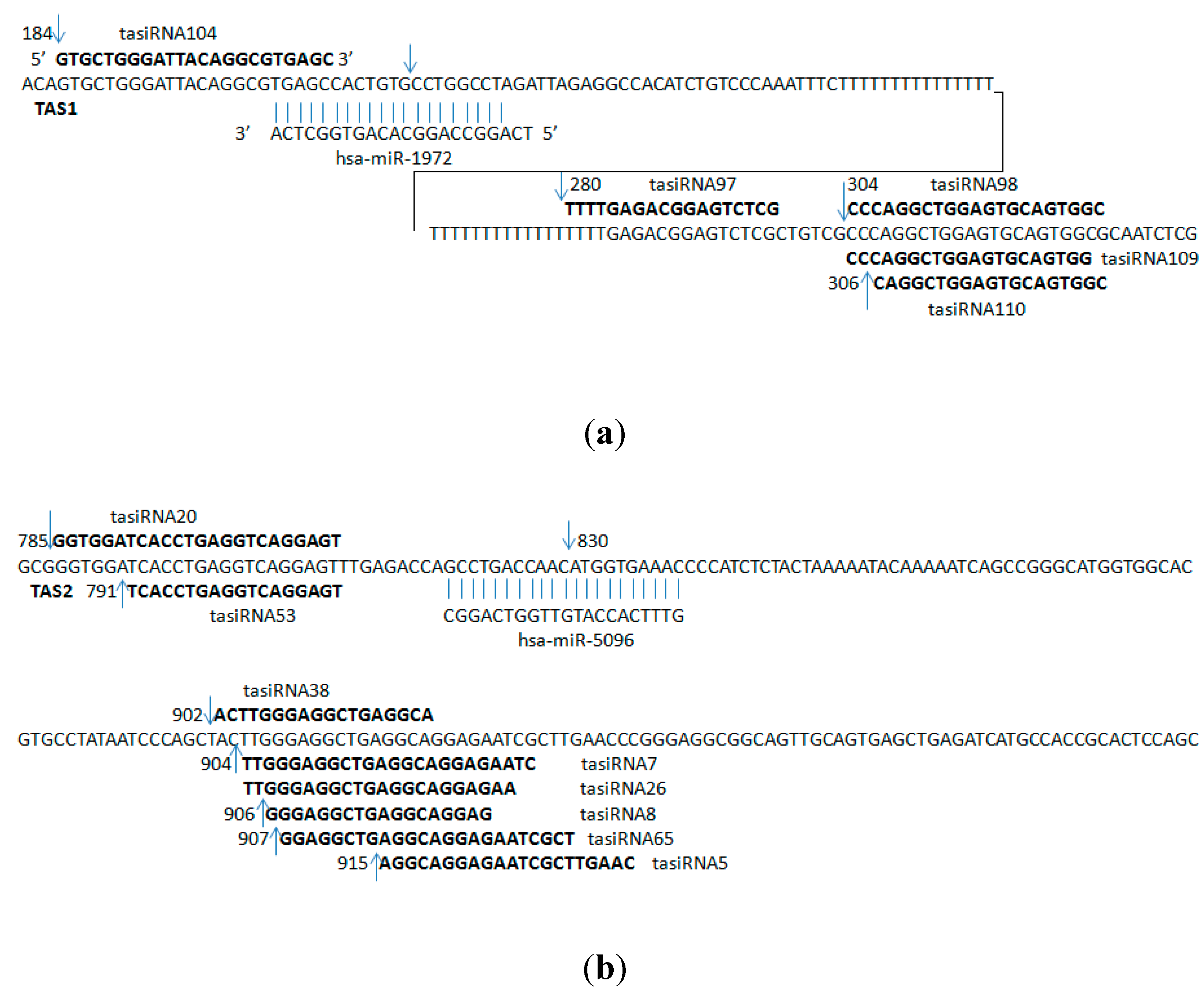
2.2. Predicted ta-siRNAs in the Human Brain
| miRNA | TAS Gene | ta-siRNA | ||
|---|---|---|---|---|
| Ensembl Transcript ID | Position | Sequence | No. of Reads | |
| hsa-miR-1972 | ENST00000547850 | 184 | GTGCTGGGATTACAGGCGTGAGC | 3 |
| ENST00000547850 | 280 | TTTTGAGACGGAGTCTCG | 2 | |
| ENST00000547850 | 304 | CCCAGGCTGGAGTGCAGTGGC | 2 | |
| ENST00000547850 | 304 | CCCAGGCTGGAGTGCAGTGG | 2 | |
| ENST00000547850 | 306 | CAGGCTGGAGTGCAGTGGC | 4 | |
| hsa-miR-5096 | ENST00000409982 | 785 | GGTGGATCACCTGAGGTCAGGAGT | 2 |
| ENST00000409982 | 791 | TCACCTGAGGTCAGGAGT | 24 | |
| ENST00000409982 | 902 | ACTTGGGAGGCTGAGGCA | 2 | |
| ENST00000409982 | 904 | TTGGGAGGCTGAGGCAGGAGAATC | 2 | |
| ENST00000409982 | 904 | TTGGGAGGCTGAGGCAGGAGAA | 2 | |
| ENST00000409982 | 906 | GGGAGGCTGAGGCAGGAG | 3 | |
| ENST00000409982 | 907 | GGAGGCTGAGGCAGGAGAATCGCT | 2 | |
| ENST00000409982 | 915 | AGGCAGGAGAATCGCTTGAAC | 2 | |
2.3. Murine ta-siRNA Prediction Analysis
2.4. Comparison of Human and Murine Predicted ta-siRNAs
| miRNA | TAS Gene | ta-siRNA | |||
|---|---|---|---|---|---|
| Ensembl Transcript ID | Description | Position | Sequence | No. of Reads | |
| hsa-miR-5095 | ENST00000338352 | OTU domain containing 6A | 1382 | ATTAGCCGGGCGTGGTGGCA | 2 |
| ENST00000338352 | OTU domain containing 6A | 1380 | AAATTAGCCGGGCGTGGTGGCA | 2 | |
| ENST00000338352 | OTU domain containing 6A | 1382 | ATTAGCCGGGCGTGGTGG | 3 | |
| ENST00000563601 | no protein product | 949 | ATTAGCCGGGCGTGGTGG | 3 | |
| ENST00000574727 | no protein product | 4185 | ATTAGCCGGGCGTGGTGG | 3 | |
| ENST00000595787 | iduronate 2-sulfatase | 5423 | ATTAGCCGGGCGTGGTGG | 3 | |
| ENST00000340855 | iduronate 2-sulfatase | 5423 | ATTAGCCGGGCGTGGTGG | 3 | |
| ENST00000470730 | no protein product | 2102 | ATTAGCCGGGCGTGGTGG | 3 | |
| hsa-miR-5585-3p | ENST00000586372 | no protein product | 2887 | ATTAGCCGGGCGTGGTGGCA | 2 |
| ENST00000586372 | no protein product | 2885 | AAATTAGCCGGGCGTGGTGGCA | 2 | |
| ENST00000586372 | no protein product | 2887 | ATTAGCCGGGCGTGGTGG | 3 | |
| ENST00000600661 | uncharacterized protein | 1644 | ATTAGCCGGGCGTGGTGG | 3 | |
| hsa-miR-1285-5p | ENST00000599386 | p21 protein (Cdc42/Rac)-activated kinase4 | 5587 | ATTAGCCGGGCGTGGTGG | 3 |
| miRNA | TAS Gene | ta-siRNA | |||
|---|---|---|---|---|---|
| Ensembl Transcript ID | Description | Position | Sequence | No. of Reads | |
| mmu-miR-1186a | ENSMUST00000151163 | no protein product | 586 | AGCCGGGCGTGGTGGCGC | 2 |
| ENSMUST00000030080 | sorting nexin family member 30 | 4190 | AGCCGGGCGTGGTGGCGC | 2 | |
| ENSMUST00000094892 | interleukin 11 | 1112 | AGCCGGGCGTGGTGGCGC | 2 | |
| mmu-miR-1186b | ENSMUST00000146029 | no protein product | 482 | AGCCGGGCGTGGTGGCGC | 2 |
| mmu-miR-3470a | ENSMUST00000125996 | no protein product | 3721 | AGCCGGGCGTGGTGGCGC | 2 |
| ENSMUST00000124097 | no protein product | 192 | AGCCGGGCGTGGTGGCGC | 2 | |
| ENSMUST00000143616 | coenzyme Q4 homolog (yeast) | 2848 | AGCCGGGCGTGGTGGCGC | 2 | |
| ENSMUST00000046525 | kringle containing transmembrane protein 2 | 1903 | AGCCGGGCGTGGTGGCGC | 2 | |
| ENSMUST00000111820 | Transmembrane protein with metallophosphoesterase domain | 3083 | AGCCGGGCGTGGTGGCGC | 2 | |
| ENSMUST00000144897 | SLX1 structure-specific endonuclease subunit homolog B (S. cerevisiae) | 1957 | AGCCGGGCGTGGTGGCGC | 2 | |
| mmu-miR-3470b | ENSMUST00000179291 | no protein product | 1868 | AGCCGGGCGTGGTGGCGC | 2 |
| ENSMUST00000168013 | MAU2 chromatid cohesion factor homolog (C. elegans) | 4087 | AGCCGGGCGTGGTGGCGC | 2 | |
| ENSMUST00000050561 | MAU2 chromatid cohesion factor homolog (C. elegans) | 4133 | AGCCGGGCGTGGTGGCGC | 2 | |
| ENSMUST00000142269 | no protein product | 4313 | AGCCGGGCGTGGTGGCGC | 2 | |
| mmu-miR-1196-5p | ENSMUST00000121443 | No protein product | 458 | AGCCGGGCGTGGTGGCGC | 2 |
| mmu-miR-3473d | ENSMUST00000098942 | SPC24, NDC80 kinetochore complex component, homolog (S. cerevisiae) | 995 | AGCCGGGCGTGGTGGCGC | 2 |
2.5. Discussion
3. Experimental Section
3.1. Datasets and Tools
3.2. Prediction Flow
3.3. Model Assessment
3.4. Conservation of ta-siRNAs between Human and Mouse
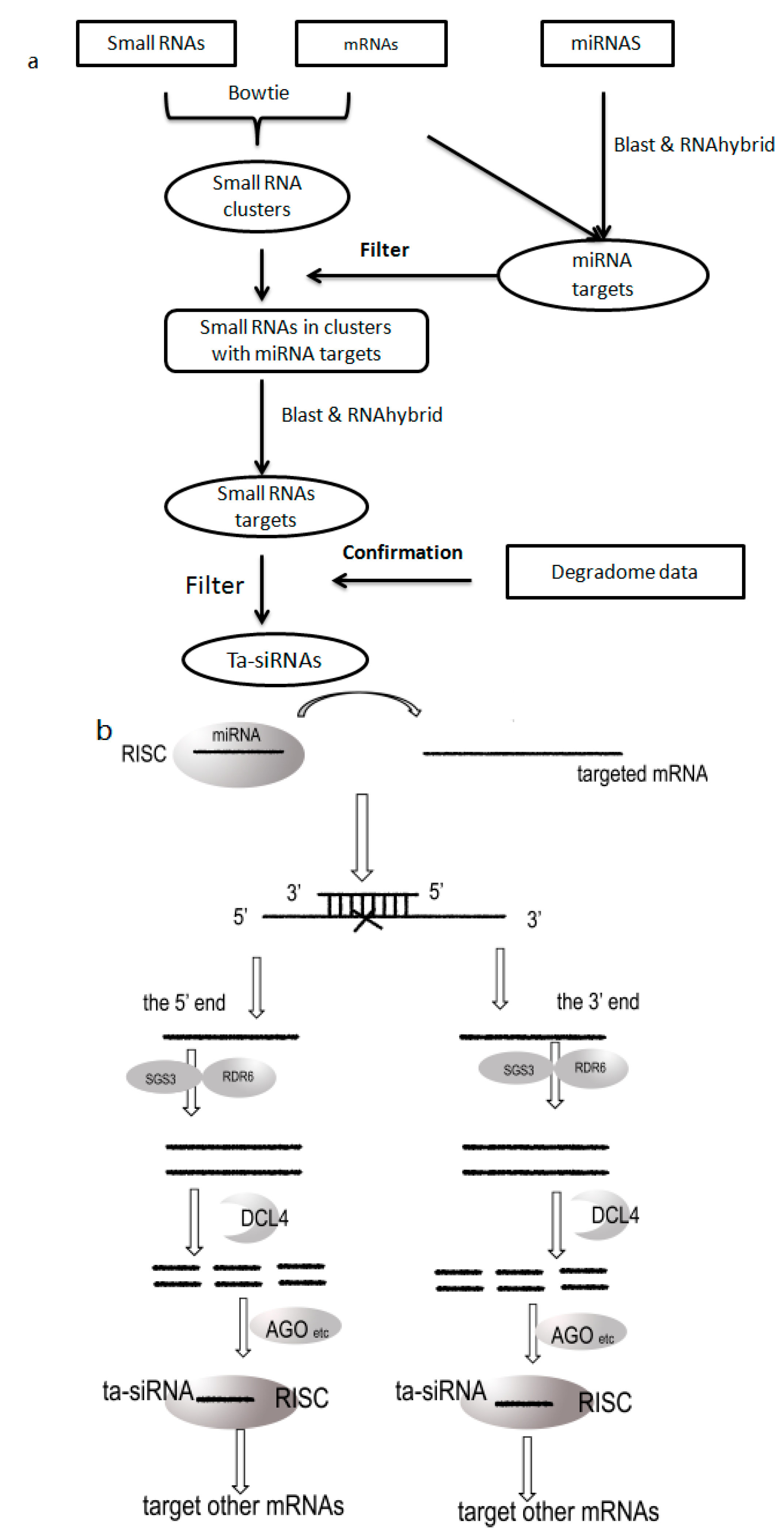
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Allen, E.; Howell, M.D. miRNAs in the biogenesis of trans-acting siRNAs in higher plants. Semin. Cell Dev. Biol. 2010, 21, 798–804. [Google Scholar] [CrossRef] [PubMed]
- Slotkin, R.K.; Martienssen, R. Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 2007, 8, 272–285. [Google Scholar] [CrossRef] [PubMed]
- Hannon, G.J. RNA interference. Nature 2002, 418, 244–251. [Google Scholar] [CrossRef] [PubMed]
- Cogoni, C.; Macino, G. Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature 1999, 399, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Dalmay, T.; Hamilton, A.; Rudd, S.; Angell, S.; Baulcombe, D.C. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 2000, 101, 543–553. [Google Scholar] [CrossRef] [PubMed]
- Mourrain, P.; Beclin, C.; Elmayan, T.; Feuerbach, F.; Godon, C.; Morel, J.B.; Jouette, D.; Lacombe, A.M.; Nikic, S.; Picault, N.; et al. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 2000, 101, 533–542. [Google Scholar] [CrossRef] [PubMed]
- Sijen, T.; Fleenor, J.; Simmer, F.; Thijssen, K.L.; Parrish, S.; Timmons, L.; Plasterk, R.H.; Fire, A. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 2001, 107, 465–476. [Google Scholar] [CrossRef] [PubMed]
- Smardon, A.; Spoerke, J.M.; Stacey, S.C.; Klein, M.E.; Mackin, N.; Maine, E.M. EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr. Biol. 2000, 10, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Peragine, A.; Yoshikawa, M.; Wu, G.; Albrecht, H.L.; Poethig, R.S. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 2004, 18, 2368–2379. [Google Scholar] [CrossRef] [PubMed]
- Vazquez, F.; Vaucheret, H.; Rajagopalan, R.; Lepers, C.; Gasciolli, V.; Mallory, A.C.; Hilbert, J.L.; Bartel, D.P.; Crete, P. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell 2004, 16, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Allen, E.; Xie, Z.; Gustafson, A.M.; Carrington, J.C. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 2005, 121, 207–221. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, M.; Peragine, A.; Park, M.Y.; Poethig, R.S. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 2005, 19, 2164–2175. [Google Scholar] [CrossRef] [PubMed]
- Axtell, M.J.; Jan, C.; Rajagopalan, R.; Bartel, D.P. A two-hit trigger for siRNA biogenesis in plants. Cell 2006, 127, 565–577. [Google Scholar] [CrossRef] [PubMed]
- Talmor-Neiman, M.; Stav, R.; Klipcan, L.; Buxdorf, K.; Baulcombe, D.C.; Arazi, T. Identification of trans-acting siRNAs in moss and an RNA-dependent RNA polymerase required for their biogenesis. Plant J. Cell Mol. Biol. 2006, 48, 511–521. [Google Scholar] [CrossRef]
- Xie, Z.; Qi, X. Diverse small RNA-directed silencing pathways in plants. Biochim. Biophys. Acta 2008, 1779, 720–724. [Google Scholar] [CrossRef] [PubMed]
- Rajagopalan, R.; Vaucheret, H.; Trejo, J.; Bartel, D.P. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006, 20, 3407–3425. [Google Scholar] [CrossRef] [PubMed]
- Mach, J. Chloroplast RNA editing by pentatricopeptide repeat proteins. Plant Cell 2009, 21. [Google Scholar] [CrossRef] [PubMed]
- Stracke, R.; Werber, M.; Weisshaar, B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001, 4, 447–456. [Google Scholar] [CrossRef] [PubMed]
- Williams, L.; Carles, C.C.; Osmont, K.S.; Fletcher, J.C. A database analysis method identifies an endogenous trans-acting short-interfering RNA that targets the Arabidopsis ARF2, ARF3, and ARF4 genes. Proc. Nat. Acad. Sci. USA 2005, 102, 9703–9708. [Google Scholar] [CrossRef] [PubMed]
- Heisel, S.E.; Zhang, Y.; Allen, E.; Guo, L.; Reynolds, T.L.; Yang, X.; Kovalic, D.; Roberts, J.K. Characterization of unique small RNA populations from rice grain. PLoS One 2008, 3, e2871. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Li, G.; Wang, J.; Fang, J. Identification of trans-acting siRNAs and their regulatory cascades in grapevine. Bioinformatics 2012, 28, 2561–2568. [Google Scholar] [CrossRef] [PubMed]
- Xia, R.; Zhu, H.; An, Y.Q.; Beers, E.P.; Liu, Z. Apple miRNAs and tasiRNAs with novel regulatory networks. Genome Biol. 2012, 13. [Google Scholar] [CrossRef]
- Zhu, H.; Xia, R.; Zhao, B.; An, Y.Q.; Dardick, C.D.; Callahan, A.M.; Liu, Z. Unique expression, processing regulation, and regulatory network of peach (Prunus persica) miRNAs. BMC Plant Biol. 2012, 12. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.M.; Li, Y.H.; Wu, S.H. Bioinformatic prediction and experimental validation of a microRNA-directed tandem trans-acting siRNA cascade in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 3318–3323. [Google Scholar] [CrossRef] [PubMed]
- Tuschl, T.; Borkhardt, A. Small interfering RNAs: A revolutionary tool for the analysis of gene function and gene therapy. Mol. Interv. 2002, 2, 158–167. [Google Scholar] [CrossRef] [PubMed]
- Howell, M.D.; Fahlgren, N.; Chapman, E.J.; Cumbie, J.S.; Sullivan, C.M.; Givan, S.A.; Kasschau, K.D.; Carrington, J.C. Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell 2007, 19, 926–942. [Google Scholar] [CrossRef] [PubMed]
- Allen, E.; Xie, Z.; Gustafson, A.M.; Sung, G.H.; Spatafora, J.W.; Carrington, J.C. Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat. Genet. 2004, 36, 1282–1290. [Google Scholar] [CrossRef] [PubMed]
- Colinas, J.; Schmidler, S.C.; Bohrer, G.; Iordanov, B.; Benfey, P.N. Intergenic and genic sequence lengths have opposite relationships with respect to gene expression. PLoS One 2008, 3, e3670. [Google Scholar] [CrossRef] [PubMed]
- Geddy, R.; Brown, G.G. Genes encoding pentatricopeptide repeat (PPR) proteins are not conserved in location in plant genomes and may be subject to diversifying selection. BMC Genomics 2007, 8. [Google Scholar] [CrossRef] [PubMed]
- Quatrini, R.; Lefimil, C.; Veloso, F.A.; Pedroso, I.; Holmes, D.S.; Jedlicki, E. Bioinformatic prediction and experimental verification of Fur-regulated genes in the extreme acidophile Acidithiobacillus ferrooxidans. Nucleic Acids Res. 2007, 35, 2153–2166. [Google Scholar] [CrossRef] [PubMed]
- Lobbes, D.; Rallapalli, G.; Schmidt, D.D.; Martin, C.; Clarke, J. SERRATE: A new player on the plant microRNA scene. EMBO Rep. 2006, 7, 1052–1058. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, T.A.; Yoo, S.J.; Fahlgren, N.; Gilbert, S.D.; Howell, M.D.; Sullivan, C.M.; Alexander, A.; Nguyen, G.; Allen, E.; Ahn, J.H.; et al. AGO1-miR173 complex initiates phased siRNA formation in plants. Proc. Natl. Acad. Sci. USA 2008, 105, 20055–20062. [Google Scholar] [CrossRef] [PubMed]
- Rajeswaran, R.; Aregger, M.; Zvereva, A.S.; Borah, B.K.; Gubaeva, E.G.; Pooggin, M.M. Sequencing of RDR6-dependent double-stranded RNAs reveals novel features of plant siRNA biogenesis. Nucleic Acids Res. 2012, 40, 6241–6254. [Google Scholar] [CrossRef] [PubMed]
- Dal Bosco, C.; Lezhneva, L.; Biehl, A.; Leister, D.; Strotmann, H.; Wanner, G.; Meurer, J. Inactivation of the chloroplast ATP synthase gamma subunit results in high non-photochemical fluorescence quenching and altered nuclear gene expression in Arabidopsis thaliana. J. Biol. Chem. 2004, 279, 1060–1069. [Google Scholar] [CrossRef] [PubMed]
- Dunoyer, P.; Himber, C.; Voinnet, O. DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nat. Genet. 2005, 37, 1356–1360. [Google Scholar] [CrossRef]
- Willmann, M.R.; Endres, M.W.; Cook, R.T.; Gregory, B.D. The Functions of RNA-Dependent RNA Polymerases in Arabidopsis. Arabidopsis Book Am. Soc. Plant Biol. 2011, 9. [Google Scholar] [CrossRef]
- Zhang, X.; Yuan, Y.R.; Pei, Y.; Lin, S.S.; Tuschl, T.; Patel, D.J.; Chua, N.H. Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 2006, 20, 3255–3268. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yu, X.; Zack, D.J.; Zhu, H.; Qian, J. TiGER: A database for tissue-specific gene expression and regulation. BMC Bioinform. 2008, 9. [Google Scholar] [CrossRef]
- Yu, X.; Lin, J.; Masuda, T.; Esumi, N.; Zack, D.J.; Qian, J. Genome-wide prediction and characterization of interactions between transcription factors in Saccharomyces cerevisiae. Nucleic Acids Res. 2006, 34, 917–927. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Lin, J.; Zack, D.J.; Qian, J. Computational analysis of tissue-specific combinatorial gene regulation: Predicting interaction between transcription factors in human tissues. Nucleic Acids Res. 2006, 34, 4925–4936. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Lin, J.; Zack, D.J.; Qian, J. Identification of tissue-specific cis-regulatory modules based on interactions between transcription factors. BMC Bioinform. 2007, 8. [Google Scholar] [CrossRef]
- Babiarz, J.E.; Ruby, J.G.; Wang, Y.; Bartel, D.P.; Blelloch, R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008, 22, 2773–2785. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Saha, D.; Prasad, A.M.; Srinivasan, R. Pentatricopeptide repeat proteins and their emerging roles in plants. Plant Physiol. Biochem. 2007, 45, 521–534. [Google Scholar] [CrossRef] [PubMed]
- Lurin, C.; Andres, C.; Aubourg, S.; Bellaoui, M.; Bitton, F.; Bruyere, C.; Caboche, M.; Debast, C.; Gualberto, J.; Hoffmann, B.; et al. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 2004, 16, 2089–2103. [Google Scholar] [CrossRef] [PubMed]
- Moran, Y.; Fredman, D.; Praher, D.; Li, X.Z.; Wee, L.M.; Rentzsch, F.; Zamore, P.D.; Technau, U.; Seitz, H. Cnidarian microRNAs frequently regulate targets by cleavage. Genome Res. 2014, 24, 651–663. [Google Scholar] [CrossRef] [PubMed]
- Alder, M.N.; Dames, S.; Gaudet, J.; Mango, S.E. Gene silencing in Caenorhabditis elegans by transitive RNA interference. RNA 2003, 9, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Lipardi, C.; Wei, Q.; Paterson, B.M. RNAi as random degradative PCR: siRNA primers convert mRNA into dsRNAs that are degraded to generate new siRNAs. Cell 2001, 107, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, E.; Brueckner, F.; Cramer, P. Molecular basis of RNA-dependent RNA polymerase II activity. Nature 2007, 450, 445–449. [Google Scholar] [CrossRef] [PubMed]
- Lipardi, C.; Paterson, B.M. Identification of an RNA-dependent RNA polymerase in Drosophila involved in RNAi and transposon suppression. Proc. Natl. Acad. Sci. USA 2009, 106, 15645–15650. [Google Scholar] [CrossRef] [PubMed]
- Maida, Y.; Yasukawa, M.; Furuuchi, M.; Lassmann, T.; Possemato, R.; Okamoto, N.; Kasim, V.; Hayashizaki, Y.; Hahn, W.C.; Masutomi, K. An RNA-dependent RNA polymerase formed by TERT and the RMRP RNA. Nature 2009, 461, 230–235. [Google Scholar] [CrossRef] [PubMed]
- Pelczar, H.; Woisard, A.; Lemaitre, J.M.; Chachou, M.; Andeol, Y. Evidence for an RNA polymerization activity in axolotl and Xenopus egg extracts. PLoS One 2010, 5, e14411. [Google Scholar] [CrossRef] [PubMed]
- Zamudio, J.R.; Kelly, T.J.; Sharp, P.A. Argonaute-bound small RNAs from promoter-proximal RNA polymerase II. Cell 2014, 156, 920–934. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.Q.; Li, G.P.; Wang, J.; Zhu, S.N.; Li, H.L. Cascading cis-cleavage on transcript from trans-acting siRNA-producing locus 3. Int. J. Mol. Sci. 2013, 14, 14689–14699. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10. [Google Scholar] [CrossRef]
- Rehmsmeier, M.; Steffen, P.; Hochsmann, M.; Giegerich, R. Fast and effective prediction of microRNA/target duplexes. RNA 2004, 10, 1507–1517. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Miranda, K.C.; Huynh, T.; Tay, Y.; Ang, Y.S.; Tam, W.L.; Thomson, A.M.; Lim, B.; Rigoutsos, I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell 2006, 126, 1203–1217. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.; Kasprzewska, A.; Tennessen, K.; Fernandes, J.; Nan, G.L.; Walbot, V.; Sundaresan, V.; Vance, V.; Bowman, L.H. Clusters and superclusters of phased small RNAs in the developing inflorescence of rice. Genome Res. 2009, 19, 1429–1440. [Google Scholar] [CrossRef] [PubMed]
- Meyers, B.C.; Tej, S.S.; Vu, T.H.; Haudenschild, C.D.; Agrawal, V.; Edberg, S.B.; Ghazal, H.; Decola, S. The use of MPSS for whole-genome transcriptional analysis in Arabidopsis. Genome Res. 2004, 14, 1641–1653. [Google Scholar] [CrossRef] [PubMed]
- Meyers, B.C.; Galbraith, D.W.; Nelson, T.; Agrawal, V. Methods for transcriptional profiling in plants. Be fruitful and replicate. Plant Physiol. 2004, 135, 637–652. [Google Scholar] [CrossRef] [PubMed]
- Ezkurdia, I.; Juan, D.; Rodriguez, J.M.; Frankish, A.; Diekhans, M.; Harrow, J.; Vazquez, J.; Valencia, A.; Tress, M.L. Multiple evidence strands suggest that there may be as few as 19,000 human protein-coding genes. Hum. Mol. Genet. 2014, 23, 5866–5878. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Zhang, G.; Zhang, C.; Wang, J. Predicted Trans-Acting siRNAs in the Human Brain. Int. J. Mol. Sci. 2015, 16, 3377-3390. https://doi.org/10.3390/ijms16023377
Liu X, Zhang G, Zhang C, Wang J. Predicted Trans-Acting siRNAs in the Human Brain. International Journal of Molecular Sciences. 2015; 16(2):3377-3390. https://doi.org/10.3390/ijms16023377
Chicago/Turabian StyleLiu, Xiaoshuang, Guangxin Zhang, Changqing Zhang, and Jin Wang. 2015. "Predicted Trans-Acting siRNAs in the Human Brain" International Journal of Molecular Sciences 16, no. 2: 3377-3390. https://doi.org/10.3390/ijms16023377