In Vitro Selection of Single-Stranded DNA Molecular Recognition Elements against S. aureus Alpha Toxin and Sensitive Detection in Human Serum
Abstract
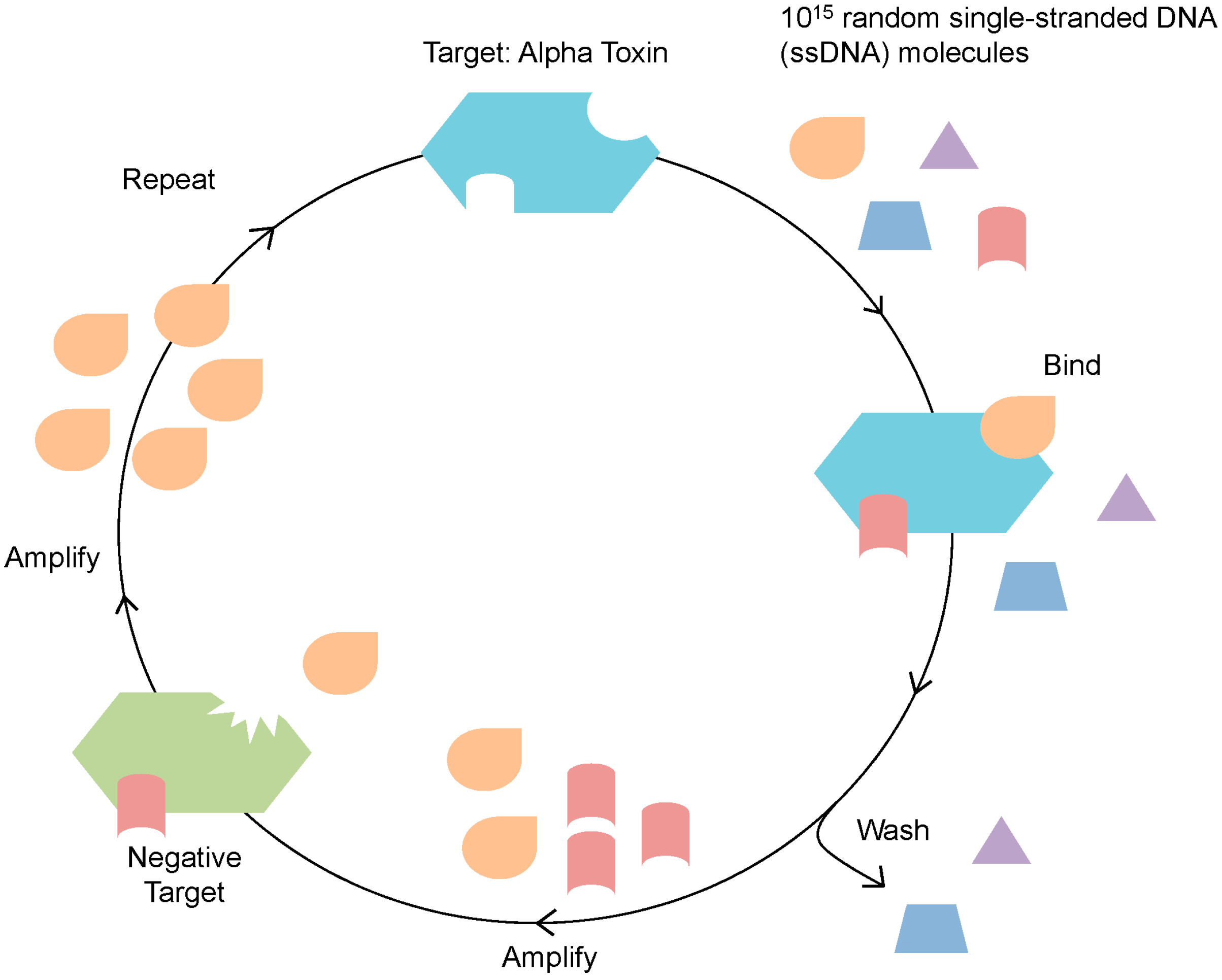
:1. Introduction
2. Results and Discussion
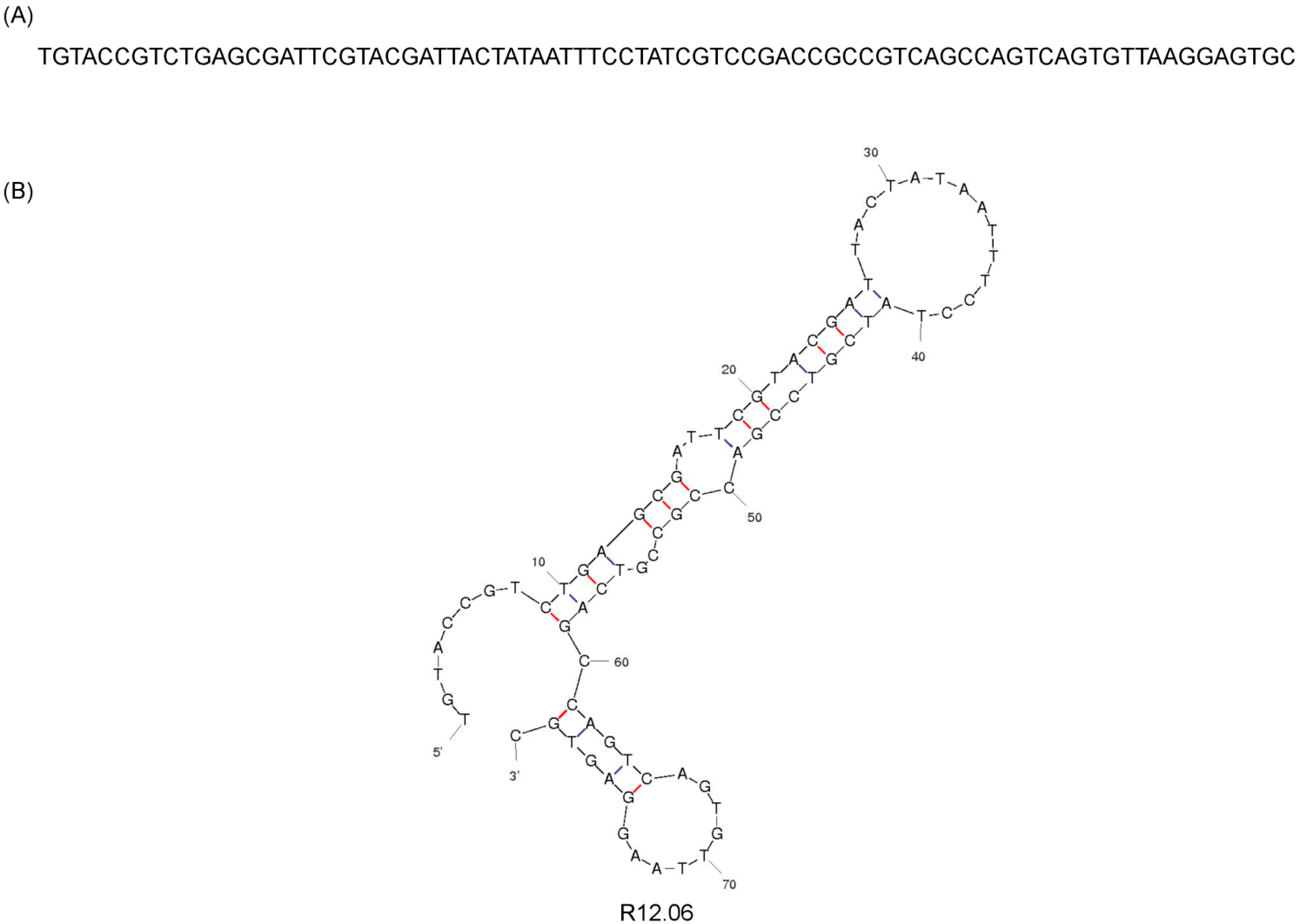
2.1. Identification of Alpha Toxin Specific MRE
| Round | Positive Selection (+) | PCR Cycles | Negative Selection (–) | PCR Cycles |
|---|---|---|---|---|
| 1 | Immobilized Target (IT) 24 h | 9 | – | – |
| 2 | IT 18 h | 15 | BSA Immobilized Negative Target (INT) 22 h | 16 |
| 3 | IT 13 h | 13 | BSA INT 26 h | 17 |
| 4 | IT 7 h | 18 | Exotoxin A INT 22 h | 16 |
| 5 | IT 3 h | 11 | Exotoxin A INT 26 h | 15 |
| 6 | IT 30 min | 17 | BSA INT 24 h | 12 |
| 7 | IT 5 min, Competitive Elution with 1 mg/mL free alpha toxin, 5 min | 17 | IT 5 min, Competitive Elution with 1 mg/mL free BSA, 5 min | 16 |
| 8 | IT 5 s, Competitive Elution with 1 mg/mL free alpha toxin, 5 s | 15 | IT 5 s, Competitive Elution with 20 µg/mL free exotoxin a, 6 h | 13 |
| 9 | IT 5 s, Competitive Elution with 10 µg/mL free alpha toxin, 5 s | 12 | IT 5 s, Competitive Elution with 20 µg/mL free cholera toxin, 6 h | 13 |
| 10 | IT 5 s, Competitive Elution with 5 µg/mL free alpha toxin, 5 s | 12 | IT 5 s, Competitive Elution with 20 µg/mL free toxin B, 6 h | 13 |
| 11 | IT 5 s, Competitive Elution with 2.5 µg/mL free alpha toxin, 5 s | 19 | IT 5 s, Competitive Elution with 20 µg/mL free BSA, 24 h | 7 |
| 12 | IT 5 s, Competitive Elution with 1 µg/mL free alpha toxin, 5 s | 10 | – | – |
| Designation Sequence |
|---|
| R12.26 TGTACCGTCTGAGCGATTCGTACCCTTGC CGATGCTCGGTAGTTGGATGTAGCCAGTCAGTGTTAAGGAGTGC |
| R12.06 TGTACCGTCTGAGCGATTCGTACGATTAAATTCTACGTCCGACCGCCGTCAGCCAGTCAGTGTTAAGGAGTGC |
| R12.01 TGTACCGTCTGAGCGATTCGTACTCGGGCGATGATACTTAGCACGGTCTAGGTCAAAAGCCAGTCAGTGTTAAGGAGTGC |
| R12.20 TGTACCGTCTGAGCGATTCGTACTAGCGGCAGAGTAGCACTCTATAGGTCGATGTTTAGCCAGTCAGTGTTAAGGAGTGC |
| R12.01 TGTACCGTCTGAGCGATTCGTACTCGGGCGATGATACTTAGCACGGTCTAG GTCAAAAGCCAGTCAGTGTTAAGGAGTGC |
| R12.02 TGTACCGTCTGAGCGATTCGTACCGGTCCTCGTTATCTCGTCAGCCAGTCAGTGTTAAGGAGTGC |
| R12.06 TGTACCGTCTGAGCGATTCGTACGATACTAAAGTCCGCGCCGTCAGCCAGTCAGTGTTAAGGAGTGC |
| R12.39 TGTACCGTCTGAGCGATTCGTACTTTGATCTCGTGTGTCTAGTTGCGGCGGATTGTCAGCCAGTCAGTGTTAAGGAGTGC |
| R12.10 TGTACCGTCTGAGCGATTCGTACGGTCAACCTCACCGACTGCCGACCGTTTAATTCGAGCCAGTCAGTGTTAAGGAGTGC |
| R12.43 TGTACCGTCTGAGCGATTCGTACCGTCATTGCCTCGTAGTATTCTTATAGTCGGTAGAGCCAGTCAGTGTTAAGGAGTGC |
| R12.44 TGTACCGTCTGAGCGATTCGTACTCCCGAAAGCGCGTCAGCCTGGGAGGTTATGCGGAGCCAGTCAGTGTTAAGGAGTGC |
2.2. Affinity and Specificity of Alpha Toxin Specific MRE
| Assay | Kd (nM) | χ2 (RU)2 |
|---|---|---|
| Assay 1 | 102 | 0.493 |
| Assay 2 | 88.7 | 0.691 |
| Assay 3 | 90.7 | 0.164 |
| Averaged | 93.7 ± 7.0 | – |
| Target | Normalized Average Fluorescence | Standard Deviation | p-Value | Selective Ratio |
|---|---|---|---|---|
| Alpha Toxin | 0.047 | 0.007 | – | – |
| Cholera Toxin | 0.031 | 0.009 | 0.003 | 1.5 |
| Exotoxin A | 0.031 | 0.002 | 0.004 | 1.5 |
| Toxin B | 0.009 | 0.002 | 0.001 | 5.0 |
| Bovine Serum Albumin | 0.026 | 0.001 | 0.027 | 1.8 |
| Human Serum | 0.028 | 0.003 | 0.017 | 1.7 |
2.3. Diagnostic Application of Alpha Toxin Specific MRE


3. Experimental Section
3.1. SELEX for Identification of Alpha Toxin Specific MREs

3.2. Cloning and Sequencing of Alpha Toxin Specific MREs
3.3. Alpha Toxin MRE SPR Affinity Binding Assays
3.4. Alpha Toxin MRE Fluorescence cross Binding Assays
3.5. Alpha Toxin MRE Modified ELISA
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bhakdi, S.; Tranum-Jensen, J. Alpha-toxin of Staphylococcus aureus. Microbiol. Rev. 1991, 55, 733–751. [Google Scholar] [PubMed]
- Lowy, F.D. Staphylococcus aureus infections. N. Engl. J. Med. 1998, 339, 520–532. [Google Scholar] [CrossRef] [PubMed]
- Mediavilla, J.R.; Chen, L.; Uhlemann, A.C.; Hanson, B.M.; Rosenthal, M.; Stanak, K.; Koll, B.; Fries, B.C.; Armellino, D.; Schilling, M.E.; et al. Methicillin-susceptible Staphylococcus aureus ST398, New York and New Jersey, USA. Emerg. Infect. Dis. 2012, 18, 700–702. [Google Scholar] [CrossRef] [PubMed]
- Vandenesch, F.; Naimi, T.; Enright, M.C.; Lina, G.; Nimmo, G.R.; Heffernan, H.; Liassine, N.; Bes, M.; Greenland, T.; Reverdy, M.E.; Etienne, J. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: Worldwide emergence. Emerg. Infect. Dis. 2003, 9, 978–984. [Google Scholar] [CrossRef] [PubMed]
- Moet, G.J.; Jones, R.N.; Biedenbach, D.J.; Stilwell, M.G.; Fritsche, T.R. Contemporary causes of skin and soft tissue infections in North America, Latin America, and Europe: Report from the SENTRY Antimicrobial Surveillance Program (1998–2004). Diagn. Microbiol. Infect. Dis. 2007, 57, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Stephens, J.M.; Gao, X.; Patel, D.A.; Verheggen, B.G.; Shelbaya, A.; Haider, S. Economic burden of inpatient and outpatient antibiotic treatment for methicillin-resistant Staphylococcus aureus complicated skin and soft-tissue infections: A comparison of linezolid, vancomycin, and daptomycin. Clinicoecon. Outcomes Res. 2013, 5, 447–457. [Google Scholar] [PubMed]
- Vandenesch, F.; Lina, G.; Henry, T. Staphylococcus aureus hemolysins, bi-component leukocidins, and cytolytic peptides: A redundant arsenal of membrane-damaging virulence factors? Front. Cell. Infect. Microbiol. 2012, 2, 12. [Google Scholar] [CrossRef] [PubMed]
- Jacobsson, G.; Colque-Navarro, P.; Gustafsson, E.; Andersson, R.; Mollby, R. Antibody responses in patients with invasive Staphylococcus aureus infections. Eur. J. Clin. Microbiol. Infect. Dis. 2010, 29, 715–725. [Google Scholar] [CrossRef] [PubMed]
- Bubeck Wardenburg, J.; Patel, R.J.; Schneewind, O. Surface proteins and exotoxins are required for the pathogenesis of Staphylococcus aureus pneumonia. Infect. Immun. 2007, 75, 1040–1044. [Google Scholar] [CrossRef] [PubMed]
- Callegan, M.C.; Engel, L.S.; Hill, J.M.; O’Callaghan, R.J. Corneal virulence of Staphylococcus aureus: Roles of alpha-toxin and protein A in pathogenesis. Infect. Immun. 1994, 62, 2478–2482. [Google Scholar] [PubMed]
- Kennedy, A.D.; Bubeck Wardenburg, J.; Gardner, D.J.; Long, D.; Whitney, A.R.; Braughton, K.R.; Schneewind, O.; DeLeo, F.R. Targeting of alpha-hemolysin by active or passive immunization decreases severity of USA300 skin infection in a mouse model. J. Infect. Dis. 2010, 202, 1050–1058. [Google Scholar] [CrossRef] [PubMed]
- Duckworth, G.J. Diagnosis and management of methicillin resistant Staphylococcus aureus infection. BMJ 1993, 307, 1049–1052. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, D.H.; Howden, B.P. Diagnosis and management of Staphylococcus aureus bacteraemia. Intern. Med. J. 2005, 35, S17–S24. [Google Scholar] [CrossRef] [PubMed]
- Roberts, S.; Chambers, S. Diagnosis and management of Staphylococcus aureus infections of the skin and soft tissue. Intern. Med. J. 2005, 35, S97–S105. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishna, U.S.; Kingston, J.J.; Harishchandra Sripathi, M.; Batra, H.V. Taguchi optimization of duplex PCR for simultaneous identification of Staphylococcus aureus and Clostridium perfringens alpha toxins. FEMS Microbiol. Lett. 2013, 340, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Uppalapati, S.R.; Kingston, J.J.; Murali, H.S.; Batra, H.V. Generation and characterization of an inter-generic bivalent alpha domain fusion protein alphaCS from Clostridium perfringens and Staphylococcus aureus for concurrent diagnosis and therapeutic applications. J. Appl. Microbiol. 2012, 113, 448–458. [Google Scholar] [CrossRef] [PubMed]
- Reddy, P.K.; Shekar, A.; Kingston, J.J.; Sripathy, M.H.; Batra, H. Evaluation of IgY capture ELISA for sensitive detection of alpha hemolysin of Staphylococcus aureus without staphylococcal protein A interference. J. Immunol. Methods 2013, 391, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Surujballi, O.P.; Fackrell, H.B. Enzyme-linked immunosorbent assay for detection of Staphylococcus aureus alpha-toxin. J. Clin. Microbiol. 1984, 19, 394–398. [Google Scholar] [PubMed]
- Bordeaux, J.; Welsh, A.; Agarwal, S.; Killiam, E.; Baquero, M.; Hanna, J.; Anagnostou, V.; Rimm, D. Antibody validation. Biotechniques 2010, 48, 197–209. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Williams, R.; Crihfield, C.; Gattu, S.; Holland, L.; Sooter, L. In vitro selection of a single-stranded DNA molecular recognition element against atrazine. Int. J. Mol. Sci. 2014, 15, 14332–14347. [Google Scholar] [CrossRef] [PubMed]
- Williams, R.M.; Kulick, A.R.; Yedlapalli, S.; Battistella, L.; Hajiran, C.J.; Sooter, L.J. In vitro selection of a single-stranded DNA molecular recognition element specific for bromacil. J. Nucleic Acids 2014, 2014, 8. [Google Scholar] [CrossRef]
- Williams, R.M.; Maher, E.; Sooter, L.J. In vitro selection of a single-stranded DNA molecular recognition element for the pesticide malathion. Comb. Chem. High Throughput Screen. 2014, 17, 694–702. [Google Scholar] [CrossRef] [PubMed]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- Gopinath, S.C.; Hayashi, K.; Kumar, P.K. Aptamer that binds to the gD protein of herpes simplex virus 1 and efficiently inhibits viral entry. J. Virol. 2012, 86, 6732–6744. [Google Scholar] [CrossRef] [PubMed]
- Imaizumi, Y.; Kasahara, Y.; Fujita, H.; Kitadume, S.; Ozaki, H.; Endoh, T.; Kuwahara, M.; Sugimoto, N. Efficacy of base-modification on target binding of small molecule DNA aptamers. J. Am. Chem. Soc. 2013, 135, 9412–9419. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, R.; Katsamba, P.S.; Nordin, H.; Pol, E.; Myszka, D.G. Analyzing a kinetic titration series using affinity biosensors. Anal. Biochem. 2006, 349, 136–147. [Google Scholar] [CrossRef] [PubMed]
- Suenaga, E.; Mizuno, H.; Penmetcha, K.K. Monitoring influenza hemagglutinin and glycan interactions using surface plasmon resonance. Biosens. Bioelectron. 2012, 32, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Ashley, J.; Li, S.F. An aptamer based surface plasmon resonance biosensor for the detection of bovine catalase in milk. Biosens. Bioelectron. 2013, 48, 126–131. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Youn, B.S.; Park, J.W.; Niazi, J.H.; Kim, Y.S.; Gu, M.B. ssDNA aptamer-based surface plasmon resonance biosensor for the detection of retinol binding protein 4 for the early diagnosis of type 2 diabetes. Anal. Chem. 2008, 80, 2867–2873. [Google Scholar] [CrossRef] [PubMed]
- Tran, D.T.; Knez, K.; Janssen, K.P.; Pollet, J.; Spasic, D.; Lammertyn, J. Selection of aptamers against Ara h 1 protein for FO-SPR biosensing of peanut allergens in food matrices. Biosens. Bioelectron. 2013, 43, 245–51. [Google Scholar] [CrossRef] [PubMed]
- Cella, L.N.; Sanchez, P.; Zhong, W.; Myung, N.V.; Chen, W.; Mulchandani, A. Nano aptasensor for protective antigen toxin of anthrax. Anal. Chem. 2010, 82, 2042–2047. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.S.; Kim, S.G.; Lahousse, M.; Park, H.Y.; Park, H.C.; Jeong, B.; Kim, J.; Kim, S.K.; Yoon, M.Y. Screening and characterization of high-affinity ssDNA aptamers against anthrax protective antigen. J. Biomol. Screen. 2011, 16, 266–271. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Chen, X.; Duan, N.; Wu, S.; Wang, Z.; Wei, X.; Wang, Y. Selection and characterization of DNA aptamers against Staphylococcus aureus enterotoxin C1. Food Chem. 2015, 166, 623–629. [Google Scholar] [CrossRef] [PubMed]
- White, R.R.; Sullenger, B.A.; Rusconi, C.P. Developing aptamers into therapeutics. J. Clin. Investig. 2000, 106, 929–934. [Google Scholar] [CrossRef] [PubMed]
- Patel, D.J.; Suri, A.K.; Jiang, F.; Jiang, L.; Fan, P.; Kumar, R.A.; Nonin, S. Structure, recognition and adaptive binding in RNA aptamer complexes. J. Mol. Biol. 1997, 272, 645–664. [Google Scholar] [CrossRef] [PubMed]
- Dayton, S.; Hashimoto, S.; Dixon, W.; Pearce, M.L. Composition of lipids in human serum and adipose tissue during prolonged feeding of a diet high in unsaturated fat. J. Lipid Res. 1966, 7, 103–111. [Google Scholar] [PubMed]
- Park, J.W.; Kallempudi, S.S.; Niazi, J.H.; Gurbuz, Y.; Youn, B.S.; Gu, M.B. Rapid and sensitive detection of Nampt (PBEF/visfatin) in human serum using an ssDNA aptamer-based capacitive biosensor. Biosens. Bioelectron. 2012, 38, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Bruno, J.G.; Kiel, J.L. Use of magnetic beads in selection and detection of biotoxin aptamers by electrochemiluminescence and enzymatic methods. Biotechniques 2002, 32, 178–180. [Google Scholar] [PubMed]
- Mairpady Shambat, S.; Haggar, A.; Vandenesch, F.; Lina, G.; van Wamel, W.J.; Arakere, G.; Svensson, M.; Norrby-Teglund, A. Levels of alpha-toxin correlate with distinct phenotypic response profiles of blood mononuclear cells and with agr background of community-associated Staphylococcus aureus isolates. PLoS One 2014, 9, e106107. [Google Scholar]
- Wang, R.E.; Wu, H.; Niu, Y.; Cai, J. Improving the stability of aptamers by chemical modification. Curr. Med. Chem. 2011, 18, 4126–4138. [Google Scholar] [CrossRef] [PubMed]
- You, K.; Lee, S.; Im, A.; Lee, S. Aptamers as functional nucleic acids: In vitro selection and biotechnological applications. Biotechnol. Bioprocess Eng. 2003, 8, 64–75. [Google Scholar] [CrossRef]
- Vivekananda, J.; Salgado, C.; Millenbaugh, N.J. DNA aptamers as a novel approach to neutralize Staphylococcus aureus alpha-toxin. Biochem. Biophys. Res. Commun. 2014, 444, 433–438. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Hirano, N.; Kaneko, J.; Kamio, Y.; Yao, M.; Tanaka, I. 2-Methyl-2,4-pentanediol induces spontaneous assembly of staphylococcal alpha-hemolysin into heptameric pore structure. Protein Sci. 2011, 20, 448–456. [Google Scholar] [CrossRef] [PubMed]
- Wedekind, J.E.; Trame, C.B.; Dorywalska, M.; Koehl, P.; Raschke, T.M.; McKee, M.; FitzGerald, D.; Collier, R.J.; McKay, D.B. Refined crystallographic structure of Pseudomonas aeruginosa exotoxin A and its implications for the molecular mechanism of toxicity. J. Mol. Biol. 2001, 314, 823–837. [Google Scholar] [CrossRef] [PubMed]
- Bujacz, A. Structures of bovine, equine and leporine serum albumin. Acta Crystallogr. Sect. D 2012, 68, 1278–1289. [Google Scholar] [CrossRef]
- O’Neal, C.J.; Jobling, M.G.; Holmes, R.K.; Hol, W.G. Structural basis for the activation of cholera toxin by human ARF6-GTP. Science 2005, 309, 1093–1096. [Google Scholar] [CrossRef] [PubMed]
- Reinert, D.J.; Jank, T.; Aktories, K.; Schulz, G.E. Structural basis for the function of Clostridium difficile toxin B. J. Mol. Biol. 2005, 351, 973–981. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hong, K.L.; Battistella, L.; Salva, A.D.; Williams, R.M.; Sooter, L.J. In Vitro Selection of Single-Stranded DNA Molecular Recognition Elements against S. aureus Alpha Toxin and Sensitive Detection in Human Serum. Int. J. Mol. Sci. 2015, 16, 2794-2809. https://doi.org/10.3390/ijms16022794
Hong KL, Battistella L, Salva AD, Williams RM, Sooter LJ. In Vitro Selection of Single-Stranded DNA Molecular Recognition Elements against S. aureus Alpha Toxin and Sensitive Detection in Human Serum. International Journal of Molecular Sciences. 2015; 16(2):2794-2809. https://doi.org/10.3390/ijms16022794
Chicago/Turabian StyleHong, Ka L., Luisa Battistella, Alysia D. Salva, Ryan M. Williams, and Letha J. Sooter. 2015. "In Vitro Selection of Single-Stranded DNA Molecular Recognition Elements against S. aureus Alpha Toxin and Sensitive Detection in Human Serum" International Journal of Molecular Sciences 16, no. 2: 2794-2809. https://doi.org/10.3390/ijms16022794





