Abstract
Mutations induced by radiation are widely used for developing new varieties of plants. To better understand the frequency and pattern of irradiation-induced chromosomal rearrangements, we irradiated the dry seeds of Chinese Spring (CS)-Dasypyrum villosum nullisomic-tetrasomic (6A/6D) addition (6V) line (2n = 44), WD14, with 60Co-γ-rays at dosages of 100, 200, and 300 Gy. The M0 and M1 generations were analyzed using Feulgen staining and non-denaturing fluorescence in situ hybridization (ND-FISH) by using oligonucleotide probes. Abnormal mitotic behavior and chromosomes with structural changes were observed in the M0 plants. In all, 39 M1 plants had structurally changed chromosomes, with the B genome showing the highest frequency of aberrations and tendency to recombine with chromosomes of the D genome. In addition, 19 M1 plants showed a variation in chromosome number. The frequency of chromosome loss was considerably higher for 6D than for the alien chromosome 6V, indicating that 6D is less stable after irradiation. Our findings suggested that the newly obtained γ-induced genetic materials might be beneficial for future wheat breeding programs and functional gene analyses.
1. Introduction
Bread wheat (Triticum aestivum L.) is one of the most important agricultural products, accounting for 20% of global human caloric intake [1]. However, artificial selection and domestication of wheat breeding programs tend to decrease the biological diversity, thereby decreasing the resistance of varieties to various biotic and abiotic stresses and leading to heavy losses in yield. Therefore, broadening the biological diversity of current wheat varieties is necessary to increase their resistance to agricultural stresses.
Dasypyrum villosum (L.) (2n = 14, VV), a species related to wheat, is resistant to biotic causal agents of wheat diseases and abiotic stress [2]. The durable powdery mildew resistance gene, Pm21, was shown to be located on the short arm of chromosome 6V (6VS) of D. villosum [3,4]. No obvious negative agronomic traits are known to be associated with this chromosome. In wheat breeding, 6VS.6AL and 1RS.1BL are considered the most widely used translocations.
Gamma irradiation is an effective way to induce mutations and to broaden crop genetic variability. Since 1956, Sears transferred gene from Aegilops umbellulata (Zhuk.) to wheat genetic background [5], genetic transfer has been widely used in wheat breeding programs and for the creation of translocations to develop novel wheat genetic resources. To date, some wheat cultivars [6,7,8] have been successfully bred by incorporating 60Co-γ-induced mutations. Many beneficial traits have been transferred from the genomes of alien species to those of wheat by 60Co-γ [5,9,10,11,12,13]. Moreover, translocations and deletions induced by gamma rays have been useful in mapping and cloning target genes [13,14]. Most previous studies [5,9,10,11,12,13] mainly focused on alien chromosomes that were studied using cytological procedures such as C–banding and genomic in situ hybridization methods. However, cytogenetic detection is generally time-consuming and labor-intensive. Given that non-denaturing florescence in situ hybridization (ND-FISH) involves the use of oligonucleotides as probes, it is a novel and efficient technique to identify chromosomes [15].
In the present study, we provide a new insight into the breakage and refusion of wheat and D. villosum chromosomes. We showed that gamma rays produced a higher frequency of breakage in B genome chromosomes that also tended to recombine with the D genome, and that chromosome 6D was less stable after irradiation than the alien chromosome 6V. Further, we obtained 58 novel 60Co-γ-induced chromosomally altered plants by using ND-FISH. These new chromosomal structural variant lines and deletion lines could be used as new genetic resources for wheat breeding.
2. Results
2.1. FISH Pattern of the WD14 Chromosomes
The CS-D. villosum nullisomic-tetrasomic (6A/6D) addition (6V) line (2n = 44) that contained four 6D chromosomes and two 6V chromosomes without 6A was selected from the CS-D. villosum addition (6V) line#3 (2n = 44) and designated asWD14. The chromosomes of WD14 were analyzed using multi-color FISH by using oligo-pTa535-1 and oligo-pSc119.2-1 as probes. These probes could distinguish the chromosomes of A, B, and D genomes. The signal patterns on each chromosome were consistent with those reported by Tang et al. [15] with the exception of the patterns on 6D. The telomeric end of 6DS in WD14 did not show any signal of pTa535-1, whereas the end of 6DS in CS showed a significant signal [15] (Figure 1).
2.2. Investigation of Mitotic Metaphase Chromosomes in M0
M0 seeds from each dosage class (100, 200, and 300 Gy) were randomly selected and analyzed using Feulgen staining and ND-FISH. Abnormal mitotic behavior and large-scale structural aberrations of chromosomes such as deletions, translocations, dicentrics and chromatin bridges were observed in the three dosage groups (Figure 2). Normal chromosomes were recorded as N chromosomes (normal chromosomes), and significant aberrations, mainly involving chromosome fragments, were recorded as A chromosomes (abnormal chromosomes). The average chromosome number per cell was calculated; the corresponding chromosome numbers for the treated groups were as follows: 100 Gy group: 2n = 43.51 (43.24N + 0.27A); 200 Gy group: 2n = 42.26 (40.93N + 1.33A); and 300 Gy group: 2n = 40.21 (35.42N + 4.79A); Table 1. As expected, higher doses of γ-irradiation resulted in an increase in the number of chromosomal aberrations (A chromosomes).
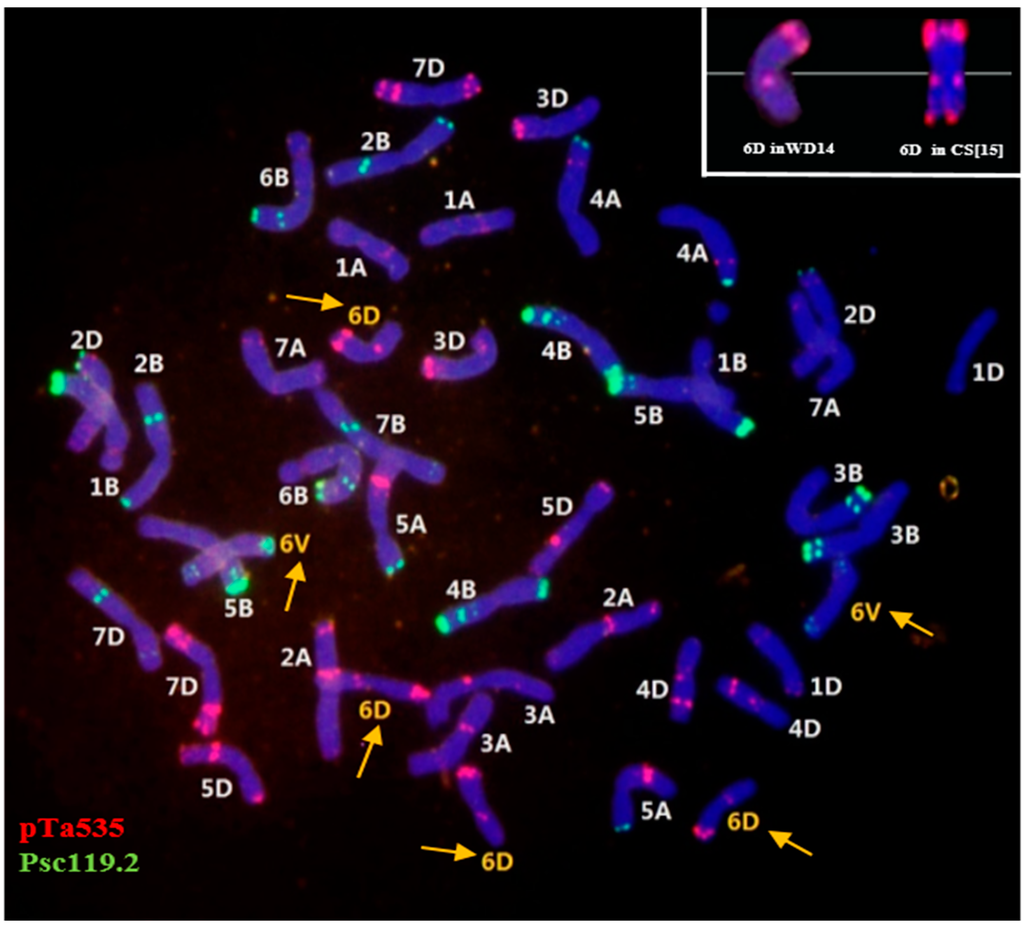
Figure 1.
Fluorescence in situ hybridization was performed on mitotic chromosomes of WD14 by using oligo-pTa535-1 (red) and oligo-pSc119.2-1 (green) as probes. Yellow arrows indicate the four 6D chromosomes and two alien chromosomes 6V from Dasypyrum villosum. Chromosomes were counterstained with 4′-6-diamidino-2-phenylindole (blue).

Table 1.
The frequency of normal chromosomes (N) and aberrations (A) in the M0 generation treated with γ-irradiation at doses of 100, 200, and 300 Gy.
| γ-Irradiation (Gy) | No. of Cells Observed | No. of Chromosomes (per cell) | Percentage of Normal Chromosomes (%) | Percentage of Abnormal Chromosomes (%) |
|---|---|---|---|---|
| 100 | 41 | 43.51 | 99.38 | 0.62 |
| 200 | 42 | 42.26 | 96.82 | 3.18 |
| 300 | 43 | 40.21 | 88.09 | 11.91 |
2.3. Chromosomal Aberrationsin M1 Plants
In all, 101 M1 plants were successfully identified using ND-FISH; these included 48 M1 plants in the 100 Gy group; 47 in the 200 Gy group; and only six in the 300 Gy group (owing to infertility). Abnormal mitotic behavior and dicentric chromosomes were not detected in the M1 plants. In contrast, chromosomal loss and rearrangements were observed in all the three dosage groups. Both wheat chromosomes and D. villosum chromosome 6V were involved in variations of chromosomal structure and number. The mutation rate increased with an increase in the radiation dose: 19 plants (39.58%) of the 100 Gy group, 27 plants (57.45%) of the 200 Gy group, and 6 plants (100%) of the 300 Gy group. Of the 101 M1 plants, 39 contained structurally changed chromosomes; these included 29 wheat-wheat translocation plants and two wheat-D. villosum translocations (4B.6V). The wheat chromosomes showing structural variations were 1A, 2A, 4A, 5A, 1B, 2B, 3B, 4B, 5B, 6B, 7B, 1D, 2D, 3D, 4D, 6D and 7D (Figure 3 and Figure S1).
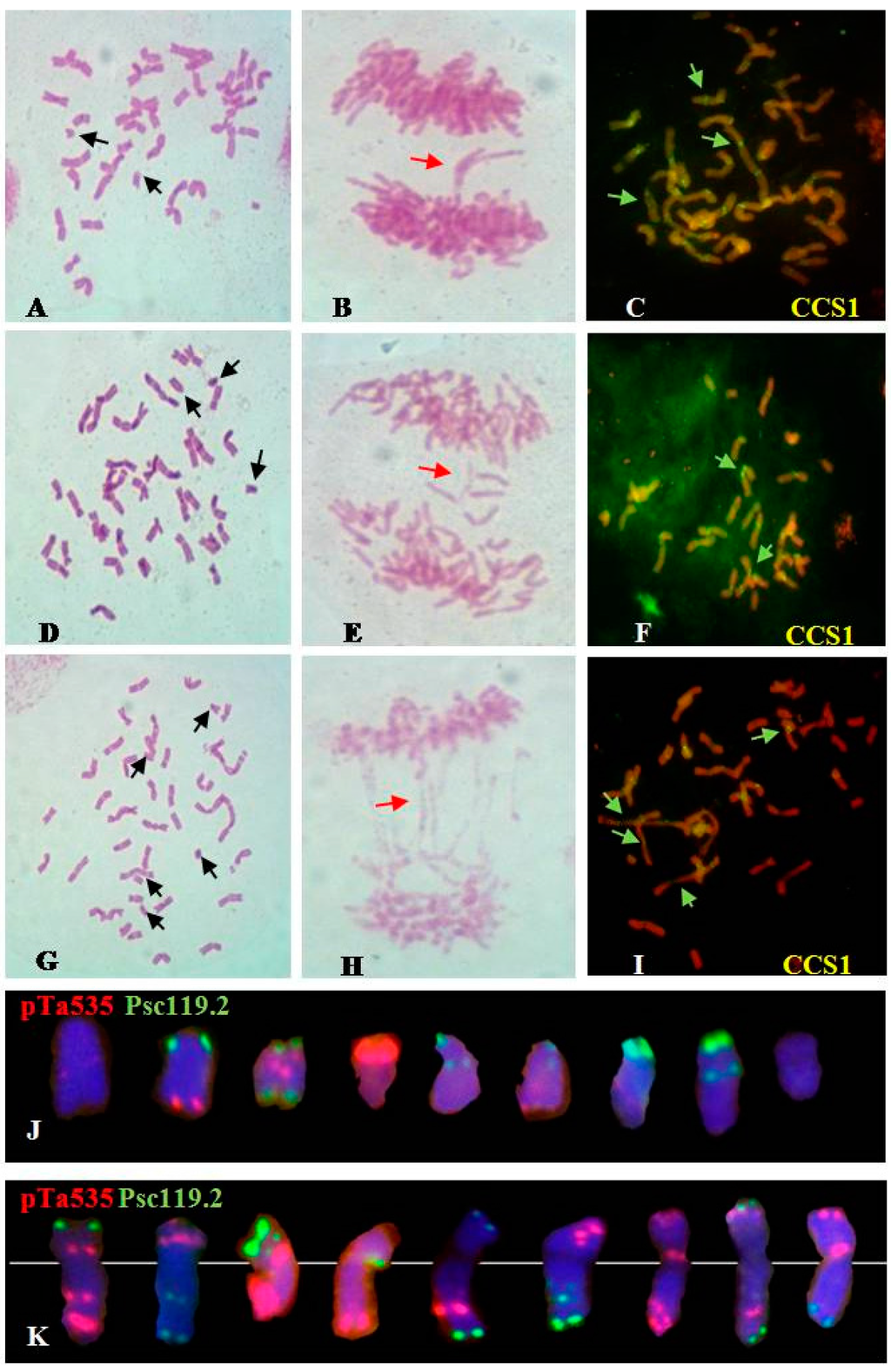
Figure 2.
Summary of the irradiation-induced chromosomal aberrations in the M0 generation. (A–C): Feulgen staining and fluorescence in situ hybridization (FISH) analyses of M0 generation of 100 Gy group; (D–F): Feulgen staining and FISH analyses of M0 generation of 200 Gy group; (G–I): Feulgen staining and FISH analyses of M0 generation of 300 Gy group; (J,K): Fragments and translocations. Black arrows indicate fragments, red arrows indicate chromatin bridges, and green arrows indicate dicentrics. Oligo-CCS1 can be used as probe to investigate centromeric structure [15]. Chromosomes were counterstained with 4′-6-diamidino-2-phenylindole (blue).
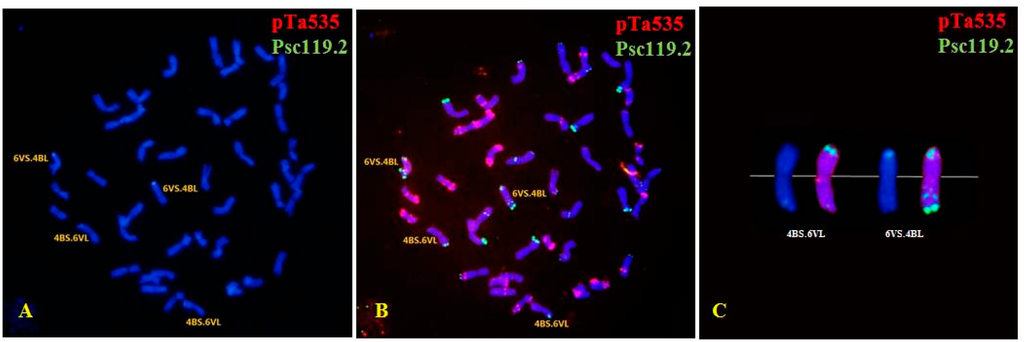
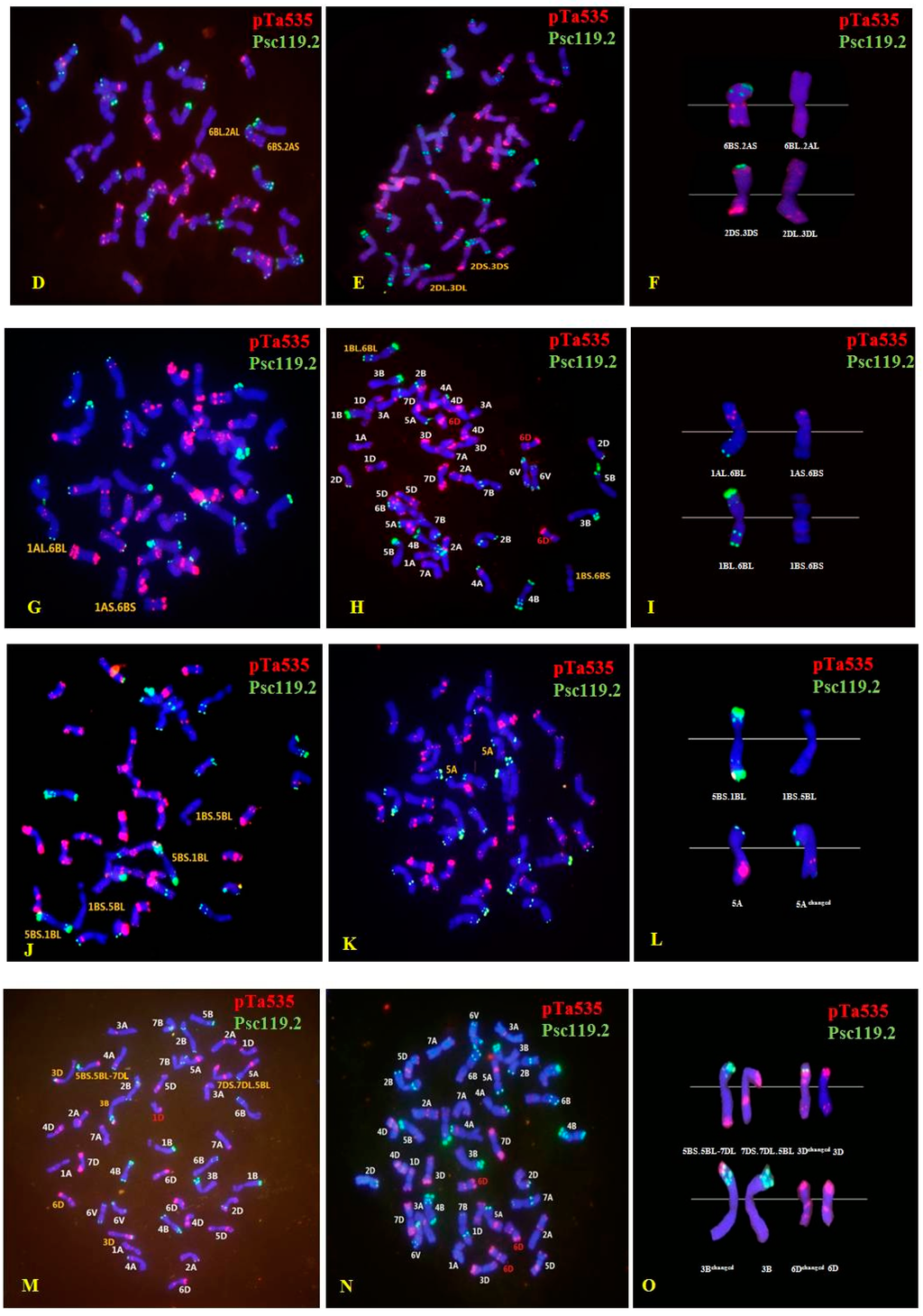
Figure 3.
Fluorescence in situ hybridization (FISH) performed usingpTa535 (red) and pSc119.2 (green) as probes for chromosomes with structural changes and mutants in the M1 generation. (A,B) 4B.6V translocation; (D) 2A.6B translocation; (E) 2D.3D translocation; (G) 1A.6B translocation; (H) 1B.6B translocation, and 6D trisome; (J) 5B.1B translocation; (K) 5A aberrations; (M) 5B.7D translocation, 3D and 6D aberrations, and 1D monosome; (N) 6D trisome. Images (C,F,I,L,O) show enlargements of the FISH pattern of chromosomes involved in the structural changes. Chromosomes were counterstained with 4′-6-diamidino-2-phenylindole (blue).
The FISH patterns of 31 plants carrying the translocation chromosomes revealed 21 interchromosomal translocation events that involved all three genomes (A, B, and D), as well as the 6V chromosome. Of the translocation events, four (19.05%) involved A and B genomes; one (4.76%), genomes A and D; 11 (52.38%), genomes B and D; two (9.52%), genome B and 6V; two (9.52%), different B genome chromosomes; and one (4.76%), different D genome chromosomes. Moreover, up to 42 breakages occurred on the chromosomes of the three wheat genomes and the 6V chromosome, including five (11.90%) breakages in the A genome, 21 (50%) in the B genome, 14 (33.33%) in the D genome and two (4.76%) on the 6V chromosome (Figure 4). These results indicated that the breakage frequency of the B genome was considerably higher than that of the A and D genomes, and that the B genome tended to recombine with the D genome.
Furthermore, other structural variations were identified. For instance, one line contained a changed 5A chromosome, whose long arm carried the weaker intercalary oligo-pTa535-1 signal (Figure 3K,L).Six lines were observed in the 300 Gy group: the 3B chromosome carrying an apparent terminal oligo-pTa535-1 signal on the short arm, a 6D chromosome with a weak terminal oligo-pTa535-1 signal on the short arm, and an extra fragment and a strong oligo-pSc119.2-1 signal on the end of 3DS (Figure 3M,O). One line carried two structurally changed 6B chromosomes without a terminaloligo-pSc119.2-1 signal on 6BL, but a strong terminal oligo-pSc119.2-1 signal on the end of the satellite (Figure S1G,I). In another plant, 4D chromosomes showed a significant terminal oligo-pSc119.2-1 signal on the long arm unlike that in the normal line (Figure S1H,I).
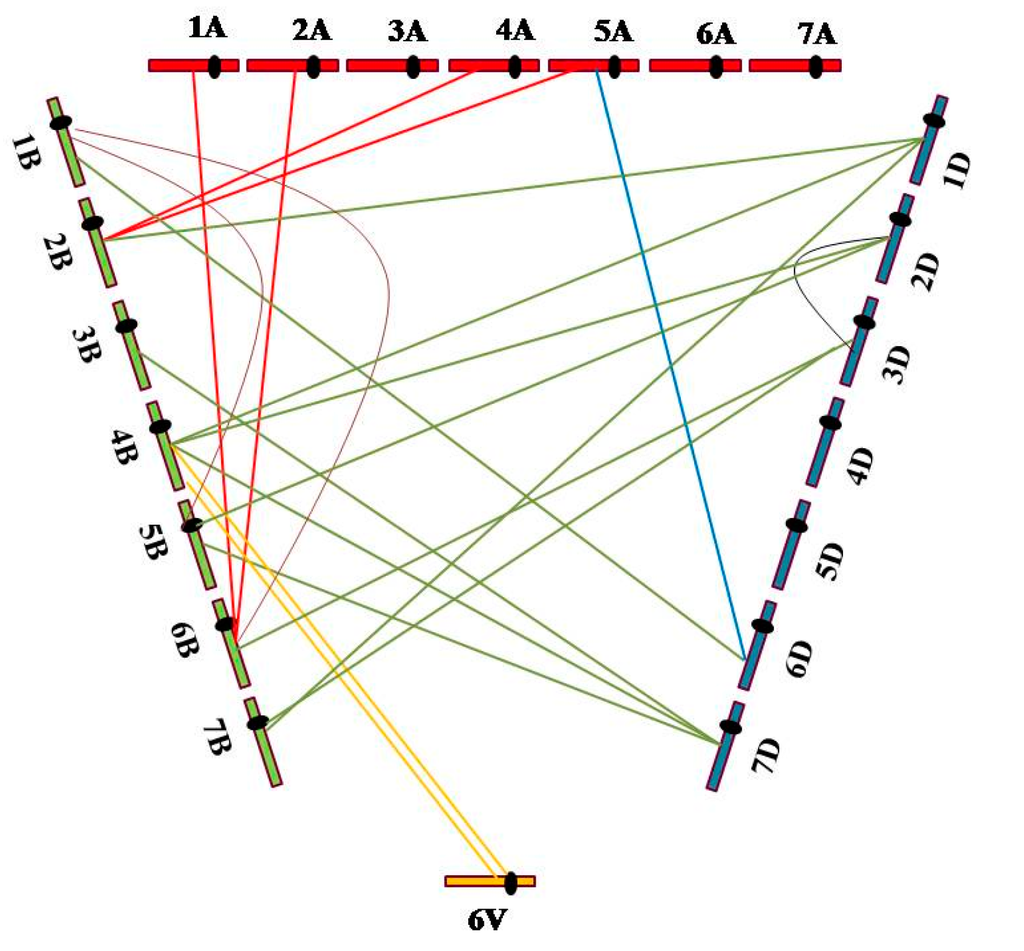
Figure 4.
Distribution of interchromosomal translocation events on chromosomes of A, B, and D wheat genomes and the alien chromosome 6V from Dasypyrum villosum.
In addition, 19 plants showing variations in chromosome number, such as nullisomic lines, monosomic lines, and trisomic lines of 6D, were also found among the 101 M1 plants. These variants involved the three wheat genomes and chromosome 6V. In total, 26 events of chromosome loss were observed in the 19 plants, among which one event involved the A genome (3.85%); nine, the B genome (34.62%); 12, the D genome (46.15%); and four, chromosome 6V (15.38%). Chromosome 6D was associated with considerably higher loss frequencies (reaching 30.77%) than the other chromosomes, whereas the alien chromosome 6V produced the second highest loss frequencies (Figure 3M,N; Figure S1D).
3. Discussion
The effects of irradiationon the frequency of chromosomal aberrations, rate of germination, and changes in morphological and physiological traits have been previously documented [16,17,18,19]. In agreement with the findings of previous studies on wheat-Aegilops biuncialis amphiploids [19,20], those of the present study suggested the presence of translocations, fragments, and dicentrics in all three dosage groups (100, 200, and 300 Gy).Furthermore, the frequency of aberrances increased with an increase in the irradiation dose in the M0 generation of the irradiated line WD14. In M1 plants, the 200 Gy group showed a higher frequency of chromosomal variation (reaching 57.45%) than those in the 100 Gy group and higher plant survival rate than that of the 300 Gy group. Therefore, we found that 200 Gy was the most optimal dosage for treating dry seeds of bread wheat.
ND-FISH is a relatively new cytogenetic technique for the rapid detection of plant chromosomes without requiring prior denaturation of chromosomes [21] by using oligonucleotides as probes [15]. Our results confirmed that ND-FISH with oligonucleotides pSc119.2-1 and pTa535-1 as probes could be used to discriminate between wheat genomes and alien chromosome 6V from D. villosum. Thirty-nine plants with structural changes in their chromosomes were detected rapidly and accurately; of these, 31 plants showed interchromosomal translocations, of which 29 were wheat-wheat and two were wheat-D. villosum. Moreover, according to FISH patterns, the B genome showed more aberrations in M1, suggesting that the B genome chromosomes are more sensitive to irradiation. Chromosomal breakpoints are known to be mainly distributed in the heterochromatin [22,23] or the border area between them [24], resulting in a higher sensitivity of the heterochromatic regions to radiation-induced breakage [25]. According to C–banding analysis, the B genome is the most heterochromatic [26]. However, larger chromosomes show aberrations more frequently than smaller ones [27], and B genome has the largest DNA content for each of the three genomes of wheat [28]. Thus, both the highest heterochromatin content and large genome of the B genome were thought to be associated with the highest sensitivity to irradiation. Ma et al. [29] detected a significantly smaller number of putative interchromosomal translocation events in the D genome and suggested that, since the A and B genomes combined considerably earlier than the D genome, the interchromosomal rearrangements seem to have occurred at a similar frequency among the A, B, and D genomes. In this study, the frequency of interchromosomal events involving the D genome were significantly higher than those of the A genome after γ-irradiation. These results indicated the presence of a moderate discrepancy between spontaneous and induced interchromosomal translocations, and the D genome was confirmed to be more sensitive to irradiation than the A genome, likely due to the high number of DNA transposons in this genome [30]. Wheat-wheat translocation lines such as 5BS.7BS have been reported to possess high resistance to powdery mildew and durable resistance to yellow rust [31,32], implying that these lines could be useful genetic blocks for wheat breeding (with the exception of wheat-alien translocations such as 1RS.1BL and 6VS.6AL). In this study, we detected 29 wheat-wheat translocation plants; some of these as well as some plants of the M1 generation possessing favorable new traits (Figure S2) might be selected and would be useful for introduction in breeding programs.
In addition to structural aberrations, 19 plants showed variations in chromosome number. As expected, FISH confirmed the events of chromosome loss in the A, B, and D genomes. According to FISH analyses, the highest frequency of chromosome loss occurred on 6D rather than on the alien chromosome 6V. The highest frequency of 6D loss might have resulted from chromosomal functional redundancy.
Taken together, our results indicated the advantage of using ND-FISH with oligonucleotides as probes to detect irradiation-induced chromosomal aberrances. The interchromosomal translocations, chromosomal structural aberrances, and deletions detected in this study are expected to be stabilized, which would become a good genetic resource to carry useful traits in wheat breeding programs as well as to map genes on chromosomes.
4. Materials and Methods
4.1. Plant Materials
Common wheat, Chinese Spring (CS)-D. villosum nullisomic-tetrasomic (6A/6D) additional (6V) line (2n = 44), WD14, was selected from CS-D. villosum additional (6V) #3 line (2n = 44) and was kindly provided by Yang Zujun (University of Electronic Science and Technology of China, Chengdu, China). Dry seeds of WD14 were subjected to 60Coγ-irradiation at dosages of 100, 200, and 300 Gy (dose rate = 1.0 Gy/min) at the Institute of Biological and Nuclear Technology, Sichuan Academy of Agricultural Sciences, Chengdu, China. The seeds treated by 60Co-γ ray were germinated, and planted in field. The mutagenized generation (M0) was analyzed using Feulgen staining and ND-FISH, and the self-pollinated progenies (M1) were analyzed using ND-FISH to identify chromosomal aberrances involving the wheat genome and 6V chromosome of D. villosum.
4.2. Mitotic Studies
The preparation of root tips, and the protocols of mitotic studies were operated according to De Tomasi [33]. Mitosis behavior was analyzed using the acetocarmine squash method. Cytological observations were performed on an Olympus CX31 microscope (Olympus, Shanghai, China).
4.3. FISH Analysis
Chromosome preparations were made from root tips according to Kang et al. [34]. After were fixed in ethanol: glacial acetic acid (3:1, v/v), the root tips were squashed on microscope slides in 45% acetic acid. The slides were frozen in liquid nitrogen for one minute, and then the cover slips were removed quickly. The oligonucleotide probes pSc119.2-1, pTa-535-1, and CCS1were synthesized according to the method described by Tang et al. [15] (Invitrogen, Shanghai, China). In situ hybridization was performed according to the method described by Han et al. [35]. Microphotographs of FISH patterns were obtained using a Leica DM2500 microscope (Leica, Shanghai, China).
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/16/12/26134/s1.
Acknowledgments
We particularly thank Hai Du, College of Agronomy and Biotechnology, Southwest University, for careful reviewing and helpful comments. We thank the Project of Innovation Ability Improvement, Sichuan Province (No.2014CXSF-001), National Natural Science Foundation of China (No.31171542), China Postdoctoral Science Foundation (No.2014M552349), and the Sichuan Wheat Breeding Community for financial support.
Author Contributions
Jie Zhang, Pu Xuan, and Zujun Yang conceived and designed the experiments; Jie Zhang, Yun Jiang, and Yuanlin Guo performed the experiments; Jie Zhang, Guangrong Li, Delin Xu, and Pu Xuan analyzed the data; Jie Zhang wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
CS: Chinese Spring; ND-FISH: non-denaturing fluorescence in situ hybridization.
References
- Brenchley, R.; Spannagl, M.; Pfeifer, M.; Barker, G.L.; D’Amore, R.; Allen, A.M.; McKenzie, N.; Kramer, M.; Kerhornou, A.; Bolser, D.; et al. Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature 2012, 491, 705–710. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.J.; Chen, P.D.; Pei, G.Z.; Wang, Y.N.; Qiu, B.X.; Wang, S.L. Transfer of Haynaldia villosa chromosomes into Triticum aestivum. In Proceedings of the 7th international wheat genet symposium, Cambridge, UK, 13–19 July 1988; Miller, T.E., Koebner, R.M.D., Eds.; Institute of Plant Science Research, Cambridge Laboratory: Cambridge, UK, 1988; pp. 355–361. [Google Scholar]
- Qi, L.L.; Wang, S.L.; Chen, P.D.; Liu, D.J.; Gill, B.S. Identification and physical mapping of three Haynaldia villosa chromosome-6V deletion lines. Theor. Appl. Genet. 1998, 97, 1042–1046. [Google Scholar] [CrossRef]
- Cao, A.Z.; Xing, L.P.; Wang, X.Y.; Yang, X.M.; Wang, W.; Sun, Y.L.; Qian, C.; Ni, J.L.; Chen, Y.P.; Liu, D.J.; et al. Serine/threonine kinase gene Stpk-V, a key member of powdery mildew resistance gene Pm21, confers powdery mildew resistance in wheat. Proc. Natl. Acad. Sci. USA 2011, 108, 7727–7732. [Google Scholar] [CrossRef] [PubMed]
- Sears, E.R. The transfer of leaf rust resistance from Aegilops umbellulata into wheat. In Genetics in Plant Breeding. Brookhaven Symposia in Biology No. 9; Brookhaven National Laboratory: Upton, NY, USA, 1956; Volume 9, pp. 1–21. [Google Scholar]
- Rachovska, G.; Rachovski, G. Winter common wheat cultivar Guinneess/1322. Field Crops Stud. 2005, 2, 187–191. [Google Scholar]
- Tomlekova, N.B. Induced mutagenesis for crop improvement in Bulgaria. Plant Mutat. Rep. 2010, 2, 4–27. [Google Scholar]
- Sakin, M.A.; Yildirim, A.; Gökmen, S. Determining some yield and quality characteristics of mutants induced from adurum wheat (Triticum durum Desf.) cultivar. Turk. J. Agric. For. 2005, 29, 61–67. [Google Scholar]
- Friebe, B.; Hatchett, J.H.; Mukai, Y.; Gill, B.S.; Sebesta, E.E. Transfer of Hessian fly resistance from rye to wheat via radiation-induced terminal and intercalary chromosomal translocations. Theor. Appl. Genet. 1991, 83, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Mukai, Y.; Friebe, B.; Hatchett, J.H.; Yamamoto, M.; Gill, B.S. Molecular cytogenetic analysis of radiation-induced wheat-rye terminal and intercalary chromosomal translocations and the detection of rye chromatin specifying resistance to Hessian fly. Chromosoma 1993, 102, 88–95. [Google Scholar] [CrossRef]
- Liu, W.X.; Chen, P.D.; Liu, D.J. Development of Tritucum aestivum–Leymus racemosus translocation lines by irradiating adult plants at meiosis. Acta Bot. Sin. 1999, 41, 463–467. [Google Scholar]
- Liu, W.X.; Chen, P.D.; Liu, D.J. Studies of the development of Triticum aestivum–Leymus racemosus translocation lines by pollen irradiation. Acta Genet. Sin. 2000, 27, 44–49. [Google Scholar] [PubMed]
- Chen, P.D.; You, C.F.; Hu, Y.; Chen, S.W.; Zhou, B.; Cao, A.Z.; Wang, X. Radiation-induced translocation with reduced Haynaldia villosa chromatin at the Pm21 locus for powdery mildew resistance in wheat. Mol. Breed. 2013, 31, 477–484. [Google Scholar] [CrossRef]
- Spielmeyer, W.; Singh, R.P.; McFadden, H.; Wellings, C.R.; Huerta-Espino, J.; Kong, X.; Appels, R.; Lagudah, E.S. Fine scale genetic and physical mapping using interstitial deletion mutants of Lr34/Yr18: A disease resistance locus effective against multiple pathogens in wheat. Theor. Appl. Genet. 2008, 116, 481–490. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.X.; Yang, Z.J.; Fu, S.L. Oligonucleotides replacing the roles of repetitive sequences pAs1, pSc119.2, pTa-535, pTa 71, CCS1, and PA WRC.1 for FISH analysis. J. Appl. Genet. 2014, 55, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Molnár, I.; Benavente, E.; Molnár-Láng, M. Detection of intergenomic chromosome rearrangements in irradiated Triticum aestivum-Aegilops biuncialis amphiploids by multicolour genomic in situ hybridization. Genome 2009, 52, 156–165. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.T.; Zhu, J.S.; Jiang, Y.; Wang, X.L.; Gu, M.X.; Wang, Y.; Kang, H.Y.; Fan, X.; Sha, L.N.; Zhang, H.Q.; et al. 100Gy 60Co γ-ray induced novel mutations in tetraploid wheat. Sci. World J. 2014. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.K.; Balyan, H.S. Induced mutations in bread wheat (Triticum aestivum L.) CV. ‘Kharchia 65’ for reduced plant height and improved grain quality traits. Adv. Biol. Res. 2009, 3, 215–221. [Google Scholar]
- Singh, B.; Datte, P.S. Gamma irradiation to improve plant vigour, grain development, and yield attributes of wheat. Radiat. Phys. Chem. 2010, 79, 139–143. [Google Scholar] [CrossRef]
- Cermeno, M.C.; Lacadena, J.R. Spatial arrangement analysis of wheat and rye genomes in triticale interphase nuclei by gamma-radiation induced chromosomal interchanges. Heredity 1983, 51, 377–381. [Google Scholar] [CrossRef]
- Ángeles, C.; Hieronim, G.; Nicolás, J. A novel, simple and rapid nondenaturing FISH (ND-FISH) technique for the detection of plant telomeres. Potential used and possible target structures detected. Chromosome Res. 2009, 17, 755–762. [Google Scholar]
- Gill, B.S. Tomato cytogenetics: A search for new frontiers. In Cytogenetics of Crop Plants; Swaminathan, M.S., Gupta, P.K., Sinha, U., Eds.; Macmillan: New Delhi, India, 1983; pp. 457–480. [Google Scholar]
- Kunzel, G.; Gecheff, K.I.; Schubert, I. Different chromosomal distribution patterns of radiation-induced interchange breakpoints in barley: First post-treatment mitosis versus viable offspring. Genome 2001, 44, 128–132. [Google Scholar] [CrossRef] [PubMed]
- Badaeva, E.D.; Dedkova, O.S.; Gay, G.; Pukhalskyi, V.A.; Zelenin, A.V.; Bernard, S.; Bernard, M. Chromosomal rearrangements in wheat: their types and distribution. Genome 2007, 50, 907–926. [Google Scholar] [PubMed]
- Puerto, S.; Marcos, R.; Ramirez, M.J.; Creus, A.; Boei, J.J.W.A.; Meijiers, M.; Natarajan, A.T.; Surrallés, J. Induction, processing and persistence of radiation-induced chromosomal aberrations involving hamster euchromatin and heterochrmatin. Mutat. Res. Gen. Toxicol. Environ. Mutagen. 2000, 469, 169–179. [Google Scholar] [CrossRef]
- Gill, B.R.; Kimber, G. Giemsa C–banding and the evolution of wheat. Proc. Natl. Acad. Sci. USA 1974, 71, 4086–4090. [Google Scholar] [CrossRef] [PubMed]
- Sachs, R.K.; Hlatky, L.R.; Trask, B. Radiation-produced chromosome aberrations: Colourful clues. Trends Genet. 2000, 16, 143–146. [Google Scholar] [CrossRef]
- Ozan, H.; Tuna, M.; Arumuganathan, K. Nonadditive changes in genome size during allopolyploidization in the wheat (Aegilops-Triticum) Group. J. Hered. 2003, 94, 260–264. [Google Scholar] [CrossRef]
- Ma, J.; Stiller, J.; Zheng, Z.; Wei, Y.M.; Zheng, Y.L.; Yan, G.J.; Doležel, J.; Liu, C.J. Putative interchromosomal rearrangements in the hexaploid wheat (Triticum aestivum L.) genotype ‘Chinese Spring’ revealed by gene locations on homoeologous chromosomes. BMC Evol. Biol. 2015, 15. [Google Scholar] [CrossRef] [PubMed]
- Mayer, K.X.; Rogers, J.; Doležel, J.; Pozniak, C.; Eversole, K.; Feuillet, C.; Gill, B.; Friebe, B.; Lukaszewski, A.J.; Sourdille, P.; et al. A chromosome based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 2014, 345, 258–287. [Google Scholar]
- Wen, Y.X.; Wu, H.S.; Zhou, W.J.; Wang, E.M.; Yu, J.; Wei, R.X. Generation of wheat intra-species translocation line showing high resistance to powdery mildew and its chromosome pattern in C–banding and in situ hybridization. Acta Genet. Sin. 1997, 24, 513–518. [Google Scholar]
- Mallard, S.; Gaudet, D.; Aldeia, A.; Abelard, C.; Besnard, A.L.; Sourdille, P.; Dedryver, F. Genetic analysis of durable resistance to yellow rust in bread wheat. Theor. Appl. Genet. 2005, 110, 1401–1409. [Google Scholar] [CrossRef] [PubMed]
- De Tomasi, J.A. Improving the technic of the feulgen stain. Biotech. Histochem. 1936, 11, 137–144. [Google Scholar] [CrossRef]
- Kang, H.Y.; Zhong, M.Y.; Xie, Q.; Zhang, H.Q.; Fan, X.; Sha, L.N.; Xu, L.L.; Zhou, Y.H. Production and cytogenetics of trigeneric hybrid involving Triticum, Psathyrostachys and Secale. Genet. Resour. Crop Evol. 2012, 59, 445–453. [Google Scholar] [CrossRef]
- Han, F.P.; Lamb, J.C.; Birchler, A. High frequency of centromere inactivation resulting in stable dicentric chromosomes of maize. Pro. Natl. Acad. Sci. USA 2006, 103, 3238–3243. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).