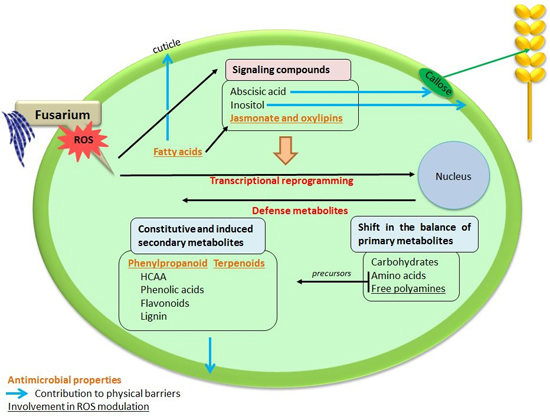
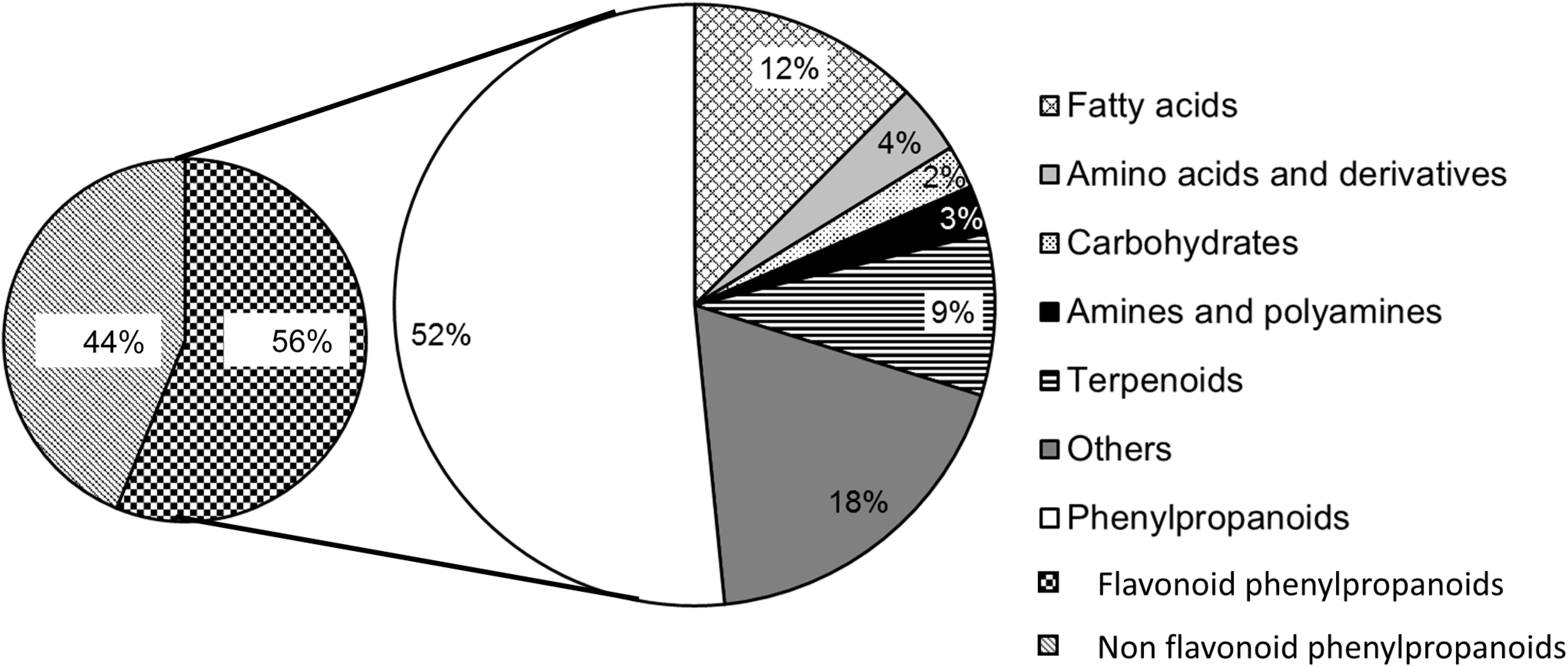
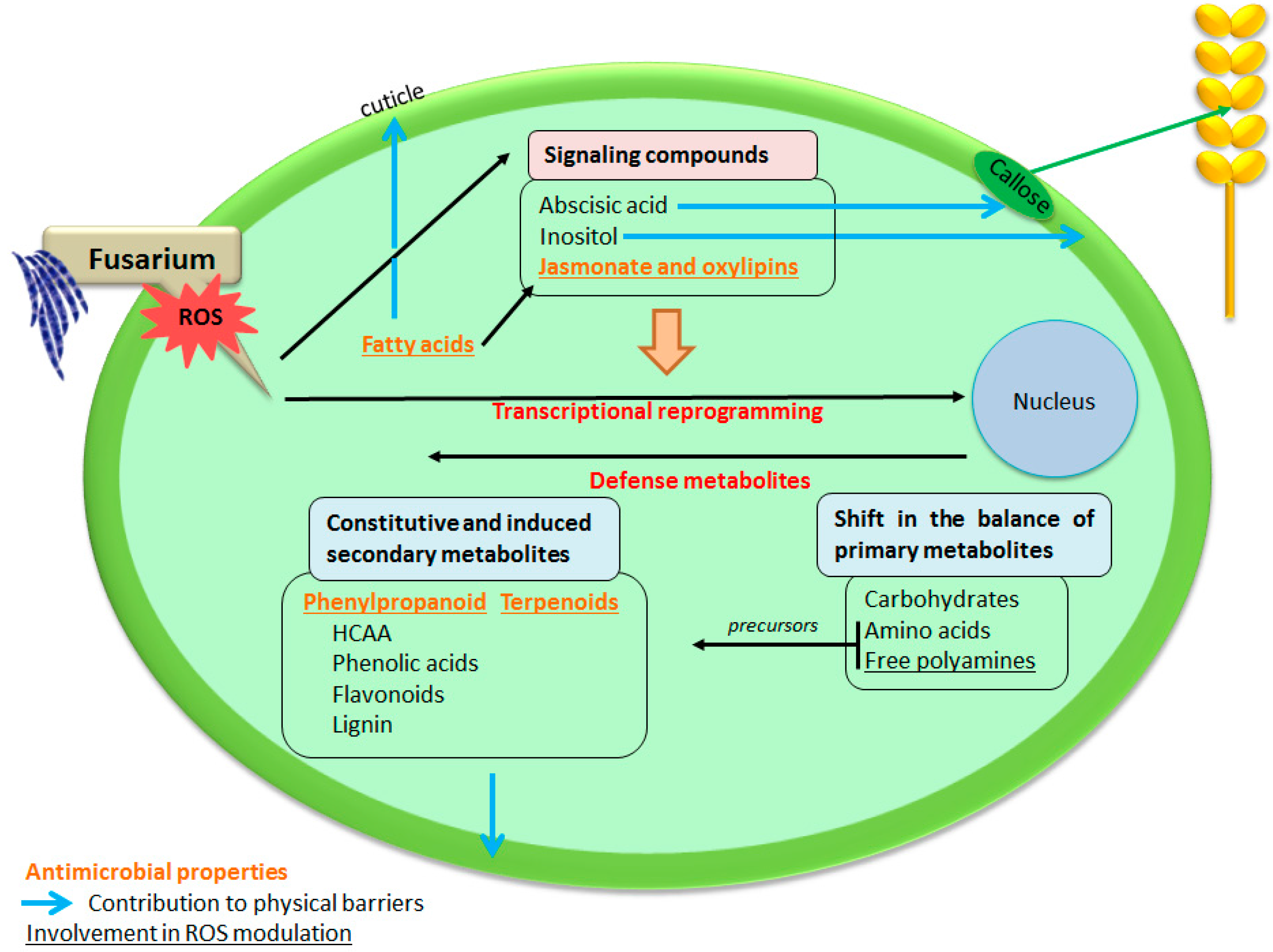
Metabolomics to Decipher the Chemical Defense of Cereals against Fusarium graminearum and Deoxynivalenol Accumulation
Abstract
:1. Introduction
2. A Large and Diverse Set of Metabolites Potentially Involved in Biochemical Resistance to FHB Spread Have Been Pinpointed through the Achievements of Metabolomic Approaches
| Pathosystem | Part of the Plant; Stage of Inoculation ab; Harvesting a | Technology | Database | Putatively Identified Metabolites Linked to FHB Resistance | Main Chemical Groups | Reference | |
|---|---|---|---|---|---|---|---|
| Plant | Pathogen | ||||||
| Wheat. 2 cultivars: 1 susceptible (Roblin); 1 resistant (Sumai3) | F. graminearum | Spikelets; Inoculation GS = 60–69 (anthesis); Harvested at 24 hai | GC-MS | NIST | 55 | Carbohydrates; Fatty acids; Phenylpropanoids | [27] |
| Wheat. 121 genotypes with different levels of FHB resistance | none | Leaf and stem; no inoculation Harvested at 14 days of growing | 1H NMR | Identification by spiking with standards | 10 | Amines; Amino acids; Carbohydrates | [22] |
| Wheat. 6 cultivars with different levels of FHB resistance | F. graminearum | Spikelets; Inoculation GS = 60–69 (anthesis); Harvested at 24 hai | GC-MS | GMD; NIST | 45 | Amino acids; Carbohydrates; Fatty acids; Organic acids; Phenylpropanoids; Polyamines | [28] |
| Wheat. 2 cultivars: 1 susceptible (Roblin); 1 resistant (Sumai3) | F. graminearum or DON | Spikelets; Inoculation GS = 65 (anthesis); Harvested at 48 hai | GC-MS | GMD; NIST | 47 | Amino acids; Carbohydrates; Fatty acids; Organic acids; Phnelypropanoids; Polyamines | [32] |
| Barley. 2 cultivars: 1 susceptible (Stander); 1 resistant (Chevron) | F. graminearum | Spikelets; Inoculation GS = 65–73; Harvested at 48 hai | LC-ESI-LTQ- Orbitrap | METLIN; PubChem; KNApSAcK; HMDB; MoTo | 47 | Amino acids; Fatty acids; Phenylpropanoids | [20] |
| Barley. 6 genotypes: 5 resistant (Chevron + 4 others); 1 susceptible (Stander) | F. graminearum | Spikelets; Inoculation GS = 71–75; Harvested at 72 hai | LC-ESI-LTQ-Orbitrap | Not cited | 130 | Fatty acids; Phenylpropanoids; Terpenoids | [21] |
| Barley. 2 genotypes: 1 susceptible; 1 resistant | F. graminearum | Spikelets; Inoculation at GS = 65–73 (anthesis to early milk stage); NA | LC-ESI-LTQ-Orbitrap | METLIN; KNApSAcK; MassBank; McGill-MD; KEGG; HMDB | 53 | Fatty acids; Phenylpropanoids | [30] |
| Barley. 6 genotypes: 1 susceptible; 5 resistant | F. graminearum | Spikelets; Inoculation at GS = 65 (anthesis to early milk stage); Harvested at 72 hai | LC-ESI-LTQ-Orbitrap | Databases not cited Identification by spiking with standards | 38 | Fatty acids; Phenylpropanoids | [31] |
| Wheat. 2 near Isogenic Lines with susceptible and resistant alleles of Fhb1 | F. graminearum | Rachis and spikelets; Inoculated at anthesis; Harvested at 72 hai | LC-ESI-LTQ-Orbitrap | METLIN; KNApSAcK; PMN; LIPIDMAPS; KEGG; McGill-MD | Rachises: 87 Spikelets: 57 | Fatty acids; Phenylpropanoids; Terpenoids | [26] |
| Black barley. 2 lines: 1 susceptible; 1 resistant | F. graminearum | Spikelets; Inoculation at GS = 65–73 (anthesis to early milk stage); Harvested at 72 hai | LC-ESI-LTQ-Orbitrap | Not cited | 74 | Amino acids; Carbohydrates; Fatty acids; Phenylpropanoids | [29] |
| Yellow barley. 2 lines: 1 susceptible; 1 resistant | Identification by spiking with standards | ||||||
| Barley. 5 lines: 1 susceptible; 4 resistant | F. graminearum | Spikelets; Inoculation at 50% anthesis; Harvested at 72 hai | LC-ESI-LTQ-Orbitrap | PlanyCyc; METLIN; KNApSAcK; KEGG | 76 | Alkaloids; Fatty acids; Hydroxycinnamic acids; Phenylpropanoids; Terpenoids | [24] |
| Barley. 9 varieties: Aktiv; Gladys; Henrike; Lilly; Radegast; Sebastian; Signora; Sladar; Tolar | F. culmorum | NA; Inoculated at the beginning of heading Harvested at 11 dai | UHPLC-Q-TOF-MS | PlanyCyc; METLIN; MassBank | 13 | Fatty acids; Phenylpropanoids | [23] |
| Wheat 2 cultivars: 1 susceptible (Roblin); 1 resistant (Sumai3) | F. graminearum | Rachis Inoculated GS = 65 (anthesis) Harvested at 3 dai | LC-ESI-LTQ-Orbitrap, | METLIN; MassBank; MS2T | 133 | Amino acids; Carbohydrates; Phenylpropanoids; Terpenoids | [25] |
3. Chemical Groups Potentially Involved in Chemical Defense against DON Producing Fusarium Species
3.1. Metabolites Derived from the Phenylpropanoid Pathway
3.1.1. Flavonoid Phenylpropanoids
| Subgroup | Putative Name of Identity | Reference |
|---|---|---|
| Anthocyanin | Cynanidin 3-O-glucoside | [21] |
| Malvidin 3-O-glucoside | [25] | |
| Pelargonidin 3-O-rutinoside | [29] | |
| Pelargonidin 3-O-sophoroside | [29] | |
| Chalcon | 2′,4′-Dihydroxy-6′-methoxychalcon | [29] |
| Chalconaringenin 2′-rhamnosyl-(1->4)-xyloside | [25] | |
| Chalconaringenin 2′-xyloside | [29] | |
| Flavanol | 7,4′-Dihydroxyflavan | [21,23] |
| 7-Hydroxy-5,4-dimethoxy-flavan | [21] | |
| Catechin 3-O-α-l-rhamnoside | [21] | |
| Catechin 5,7,3′-trimethyl ether | [20] | |
| Catechin 7-O-apiofuranoside | [21] | |
| Catechin | [29,30] | |
| Catechin-4-ol 3-O-β-d-galactopyranoside | [20] | |
| Catechol glucoside | [21] | |
| Epicatechin-3-O-(3-O-methylgallate) | [25] | |
| Epicatechin 5-O-β-d-glucopyranoside-3-benzoate | [21] | |
| Epigallocatechin | [25] | |
| Gallocatechin-4β-ol | [21] | |
| Flavanone | 5-Hydroxy-7,8-dimethoxyflavanone 5-rhamnoside | [26] |
| 5-Hydroxy-7,4′-dimethoxy-6,8-di-c-prenylflavanone 5-O-galactoside | [25] | |
| 5-O-Methylleridol | [21] | |
| 5′-Prenylhomoeriodictyol | [25] | |
| 6-Prenylnaringenin | [21] | |
| Dalpanin | [24] | |
| Exiguaflavanone | [31] | |
| Kuwanon L | [31] | |
| Mucronulatol-(4->6) naringenin | [21] | |
| Nallaflavanone | [1] | |
| Naringenin 7,4′-dimethyl ether | [29] | |
| Naringenin 7-O-(2′′,6′′-di-O-α-rhamnopyranosyl)-β-glucopyranoside | [25] | |
| Naringenin | [29] | |
| Naringenin-7-O-glucoside | [29,30] | |
| Naringenin 5,7-dimethyl ether 4′-O-xylosyl-(1->4)-arabinoside | [21] | |
| Tetrahydroxy-6,8-di-C-prenylflavanone | [29] | |
| 3,5,7′-Trihydroxy-4-methoxyflavone | [21,30] | |
| 5,6-Dimethoxyflavone | [26] | |
| 5,4′-Dihydroxy-3,6,7,8,2′-pentamethoxyflavone | [21,30] | |
| 5,7,2′-Trihydroxy-8,6′-dimethoxyflavone | [21,30] | |
| 5,7,3′,5′-Tetrahydroxy-8,4′-dimethoxyflavone | [31] | |
| 5-Hydroxy-3,6,7,4′-tetramethoxyflavone | [21] | |
| 6-Prenylapigenin | [21] | |
| Alpinumisoflavone | [30] | |
| Alpinumisoflavone dimethyl ether | [21] | |
| Apigenin | [21,23] | |
| Apigenin 7-O-rutinoside | [25] | |
| Apigenin 7-O-β-d-glucuronide | [21] | |
| Calophyllolide | [31] | |
| Isoorientin 4′-O-glucoside-2′′-O-(E)-caffeate | [25] | |
| Isoscoparin | [21] | |
| Isovitexin 2′′-O-(6′′′-feruloyl) glucoside | [25] | |
| Isovitexin-7-O-glucosyl-2′′-O-rhamnoside | [24] | |
| Isovitexin-7-O-xyloside | [24] | |
| Lupinisoflavone G | [25] | |
| Skullcapflavone I 2′-(4′-E-cinnamoylglucoside) | [21] | |
| Tangeretin | [21] | |
| Tetrahydroxy-6,8-di-C-prenylflavone | [29] | |
| Tricin 7-rutinoside | [21] | |
| Ulexone B | [25] | |
| Vitexin | [29] | |
| Vitexin 2′′-O-(E)-ferulate | [21] | |
| Flavonol | 3,7-Di-O-methylquercetin | [29] |
| 6-Hydroxykaempferol 7,4′-dimethyl ether 3-sulfate | [31] | |
| Dihydroquercetin | [29] | |
| Isoscoparin 7-O-glucoside | [21] | |
| Kaemferide 5-glucoside-7-glucuronide | [31] | |
| Kaempferide 3,7-diglucoside | [31] | |
| Kaempferide 3-glucoside-7-rhamnoside | [20,21] | |
| Kaempferol 3-(2′′,3′′-diacetyl-4′′-p-coumaroyl rhamnoside) | [31] | |
| Kaempferol 3-(2′′-(Z)-p-coumaroyl rhamnoside) | [20] | |
| Kaempferol 3-(6′′-caffeoyl glucoside) | [20] | |
| Kaempferol 3-apiosyl-(1->4)-rhamnoside-7-rhamnoside | [21] | |
| Kaempferol-3-gentiobioside-7-rhamnoside | [20] | |
| Kaempferol-3-glucoside-7-rhamnoside | [21] | |
| Kaempferol-3-isorhamnoside | [31] | |
| Kaempferol-3-O-glucoside 7-O-rhamnoside | [21] | |
| Kaempferol-3-rhamnoside-7-glucosyl-(1->2)-rhamnoside | [21] | |
| Kaempferol-3-rhamnoside-7-xylosyl-(1->2)-rhamnoside | [20] | |
| Kaempferol-3-sophoroside-7-rhamnoside | [20] | |
| Kaempferol-3-xyloside | [21] | |
| Kaempferol-4′-methyl ether 3-neohesperioside | [31] | |
| Kaempferol-7,4′-dirhamnoside | [21,31] | |
| Kaempferol-3-O-α-rhamnoside | [20,21,23,25] | |
| Kaempferol-3-O-α-rhamnosyl glucoside | [21] | |
| Quercetin 3,7-dimethyl ether | [21,26] | |
| Quercetin 3-(6′′-acetylglucoside) | [31] | |
| Quercetin 3-O-methyl-7-O-galactoside | [21] | |
| Quercetin 5,7,3′,4′-tetramethyl ether | [30] | |
| Quercetin 7,3′,4′-trimethyl ether | [25] | |
| Quercetin | [30] | |
| Quercetin pentamethyl ether | [21] | |
| Quercetol | [29,30] | |
| Rhamnetin 3-O-rhamninoside | [21] | |
| Isoflavone | 2-Hydroxyisoflavone naringenin | [26] |
| 7-Hydroxy-4′-methoxyisoflavone | [31] | |
| 7-Prenyloxy-3′,4′-dimethoxyisoflavone | [31] | |
| Isoflavanone | (+−)-5-Deoxykievitone | [21] |
| Sappanone A trimethyl ether | [21] | |
| Isoflavonol | Methylophiopogonone B | [25] |
3.1.2. Non Flavonoid Phenylpropanoids: Phenolic Acids and Derivatives
| Subgroup | Putative Name of Identity | Reference |
|---|---|---|
| Phenolic acid and derivatives | 1-O-Sinapoyl-β-d-glucose | [24] |
| 5-O-Feruloylquinic acid | [21] | |
| 6′-O-(p-Coumaroyl)-procumbide | [25] | |
| 7-O-(4-Methoxycinnamoyl) tecomoside | [21] | |
| 7-O-Glucoside-ferulic acid | [21] | |
| Anisic acid | [32] | |
| Benzene acetic acid | [32] | |
| Benzoic acid | [32] | |
| Caffeic acid glycoside | [25] | |
| Chlorogenic acid | [21] | |
| Cinnamic acid | [29,32] | |
| Feruloyl-3-(arabinosylxylose), cis-p-coumaric acid 4-[apiosyl-(1->2)-glucoside] | [25] | |
| Cyano-p-hydroxycinnamic acid | [29] | |
| Diferulic acid | [25] | |
| Ferulic acid | [26] | |
| Geranyl cinnamic acid | [25] | |
| Methyl 6-O-p-trans-coumaroyl-β-d-glucopyranoside | [21] | |
| Methyl cinnamic acid | [30] | |
| m-Hydroxycinnamic acid | [27] | |
| p-Coumaric acid | [21,29,30] | |
| p-Coumaroylquinic acid | [26] | |
| p-Coumaroylshikimic acid | [24] | |
| p-Methoxycinnamic acid | [21] | |
| Rosmarinic acid | [30] | |
| Salvianolic acid | [31] | |
| Sinapic acid | [21,26] | |
| α-Cyano-p-hydroxycinnamic acid | [30] | |
| β-d-Glucopyranosyl-caffeic acid | [26] | |
| β-d-Glucopyranosyl-sinapic acid | [21] | |
| Phenolic alcohol | 1-β-(3-Hydroxy-4,5-dimethoxyphenyl)-O-glucopyranoside | [21] |
| 3,4-Dihydroxystyrene | [29] | |
| Caffeoyl alcohol | [30,31] | |
| Catechol | [29] | |
| Coniferin | [21,25,29] | |
| Coniferyl alcohol | [26] | |
| Dihydroconiferyl alcohol glucoside | [24] | |
| Dihydrodiconiferoyl alcohol | [21] | |
| Gingerol | [30] | |
| Guaiacol | [29] | |
| p-Coumaryl alcohol | [29] | |
| p-Hydroxycinnamyl alcohol 4-d-glucoside | [25] | |
| Pyrogallol | [21] | |
| Sinapoyl alcohol | [25,29] | |
| Syringin | [24,26] | |
| trans-p-Ferulyl alcohol 4-O-[6,[6-(2-methyl-3-hydroxypropinyl)] glucopyranoside | [21,29] | |
| Phenolic aldehyde | Coniferaldehyde | [29] |
| Caffeyl aldehyde | [29] | |
| Sinapaldehyde | [25,26,29] | |
| Sinapaldehyde glucoside | [21] | |
| Lignan and stilbene | (+)-Pinoresinol 4-O-(6-O-galloyl)-β-d-glucopyranoside | [25] |
| (+)-Pinoresinol | [25] | |
| (+)-Syringaresinol-O-β-d-glucoside | [21] | |
| 1-Acetoxypinoresinol | [21] | |
| 3,4′,5-Trihydroxystilbene 4′-O-β-d-(6′′-O-galloyl) glucopyranoside | [31] | |
| 4′-Demethylpodophyllotoxine | [30] | |
| 4′-Prenyloxyresveratrol | [21] | |
| 5-Methoxypodophyllotoxin | [30] | |
| 6-Methoxypodophyllotoxin | [24] | |
| Astringin | [21] | |
| Bisosthenon | [31] | |
| Deoxypodophyllotoxin | [21] | |
| Diphyllin | [30] | |
| Hydnocarpin | [31] | |
| Matairesinol | [21] | |
| Medioresinol 4′-O-β-d-glucopyranoside | [21] | |
| Oxyresveratrol 2-O-β-glucopyranoside | [30] | |
| Phyllanthusmin B | [21] | |
| Podorhizol β-d-glucoside | [21,31] | |
| Secoisolariciresinol di-O-glucoside | [21] | |
| Threo-carolignan E | [25] | |
| Tuberculatin | [31] | |
| Coumarin | 4-Geranyloxy-5-methyl coumarin | [31] |
| 5-Methoxyfuranocoumarine | [29] | |
| 6,7-Dihydroxy-5-methoxycoumarin 6-β-d-glucopyranosyde | [21] | |
| Coumarin | [29,32] | |
| Dimethoxy 4-phenlycoumarin | [30] | |
| trans-Grandmarin isovalerate | [21] |
3.1.3. Non-Flavonoid Phenylpropanoids: Lignins and Lignans
3.2. Metabolites Derived from the Fatty Acid Pathway
| Putative Name of Identity | Reference |
|---|---|
| (−) Jasmonic methyl ester | [24] |
| (+)-7-iso-Jasmonoyl-l-isoleucine | [25,26] |
| (E)-Dodec-2-enedioic acid | [24,30,31] |
| 10,16-Dihydroxy-hexadecanoic acid | [20,31] |
| 10-Methyl-lauric acid | [30] |
| 12-oxo-cis-10,15-Phytodienoic acid | [24] |
| 13(S)-Hydroperoxy linolenic acid | [21,26] |
| 13(S)-Hydroperoxy-9(Z),11(E),15(Z)-octadecatrienoic acid | [24] |
| 18-oxo-Oleic acid | [24] |
| 2(Z),4(E)-Decadienoic acid | [30] |
| 2,3-Dinor-8-iso-prostaglandin F1α | [24] |
| 2,3-Dinor-8-iso-prostaglandin F2α | [24] |
| 2-Hydroxy palmitic acid | [31] |
| 2-Methoxy-6(Z)-haxadecenoic acid | [30] |
| 2-Nonadecenoic acid | [31] |
| 3-Hydroxy-3-methylglutaric acid | [21] |
| 3-Hydroxytetradecanedioic acid | [30] |
| 3-oxo-2-(2-Pentenyl)-cyclopentaneoctanoic acid | [21,24] |
| 5-Pyrophosphate mevalonic acid | [21] |
| 7-Dehydrologanin tetra acetic acid | [24] |
| 8-oxo-9,11-Octadecadiynoic acid | [21,29] |
| 9,10-Epoxy-18-hydroxystearic acid | [24] |
| 9-oxo-Nanoic acid | [24] |
| 9(S)-Hydroxy-10(E),12(Z)-octadecadienoic acid | [26] |
| Acide-5,7-nonadienoique | [29] |
| Adipate | [29] |
| Decenedioic acid | [29,30] |
| Dihydroxylinoleic acid | [29] |
| Dioxo-decanoic | [28] |
| Dodecanedioic acid | [30] |
| Dodecanoic acid | [30] |
| Eicosanoic acid | [21,24] |
| Glycerol | [21] |
| Heptadecanoic acid | [21,29] |
| Heptadetrienoic acid | [31] |
| Heptenoic acid | [30] |
| Hexadecanoic acid | [24] |
| Jasmonic acid | [21,23,26,30,31] |
| Jasmonoyl-valine | [25,26] |
| Linoleic acida | [24,30,31] |
| Linolenic acid | [21,24,26,30,31] |
| Octadeacnoic acid | [1] |
| Oleic acid | [21,30,31] |
| ω-Hydroxydodecanoic acid | [20] |
| Tetradecanoic acid | [30] |
| Tuberonic acid glucoside | [24] |
| Undecanoic acid | [20,21] |
3.2.1. Oleic, Linoleic and Linolenic Acid
3.2.2. Oxidation Products of Fatty Acids
3.2.3. The Signaling Molecules, Jasmonic Acid and Its Derivatives
3.3. Terpenoids
| Subgroup | Putative Name of Identity | Reference |
|---|---|---|
| Abscisic acid derivatives and precursors | 8′-Hydroxyabscisate | [26] |
| Abscisic alcohol | [26] | |
| Abscisic aldehyde | [26] | |
| Abscisic acid glucose ester | [26] | |
| Xanthoxin | [26,29] | |
| Iridoids and derivatives | 10-Hydroxyloganin | [26] |
| 7-Deoxyloganic acid | [26] | |
| Asperuloside | [26] | |
| Aucubin | [21,26] | |
| Deutzioside | [26] | |
| Ipolamiide | [24] | |
| Iridotrial glucoside | [24] | |
| Loganin | [26] | |
| Secologanin | [24] | |
| Hemiterpene | Isovaleroyloxylinalool | [21] |
| Diterpene | Phytocassane B | [21] |
| 16-Diacetoxy-7a-hydroxy-18-malonyloxyent-cleroda-3-ene | [21] | |
| Briaexcavatin O | [21] | |
| Brusatol | [29] | |
| Hallactone B | [24] | |
| Inumakilactone A glucoside | [24] | |
| Isobrucein A | [24] | |
| trans-Communic acid | [24] | |
| Triterpene | Cineracipadesin F | [24] |
| Quinovic acid | [21] | |
| Salannin | [29] | |
| Sesquiterpene | 2-oxo-6-Dehydroxyneoanisatin | [24] |
| 3-Hydroxy-15-dihydrolubimin | [24] | |
| Cryptomeridiol | [21] | |
| Eupacunolin | [24] | |
| Tetraterpene | Apo-13-zeaxanthione | [30] |
3.4. Amino Acids and Derivatives
3.5. Amine and Polyamine Pathway Metabolites
3.6. Carbohydrate Pathway Metabolites
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Goswami, R.S.; Kistler, H.C. Heading for disaster: Fusarium graminearum on cereal crops. Mol. Plant Pathol. 2004, 5, 515–525. [Google Scholar] [CrossRef] [PubMed]
- Bottalico, A.; Perrone, G. Toxigenic Fusarium species and mycotoxins associated with head blight in small-grain cereals in europe. Eur. J. Plant Pathol. 2002, 108, 611–624. [Google Scholar] [CrossRef]
- Desjardins, A.E. Fusarium Mycotoxins Chemistry, Genetics and Biology; APS Press: St. Paul, MN, USA, 2006. [Google Scholar]
- Hazel, C.M.; Patel, S. Influence of processing on trichothecene levels. Toxicol. Lett. 2004, 153, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Pirgozliev, S.R.; Edwards, S.G.; Hare, M.C.; Jenkinson, P. Strategies for the control of Fusarium head blight in cereals. Eur. J. Plant Pathol. 2003, 109, 731–742. [Google Scholar] [CrossRef]
- Blandino, M.; Haidukowski, M.; Pascale, M.; Plizzari, L.; Scudellari, D.; Reyneri, A. Integrated strategies for the control of Fusarium head blight and deoxynivalenol contamination in winter wheat. Field Crop. Res. 2012, 133, 139–149. [Google Scholar] [CrossRef]
- Terzi, V.; Tumino, G.; Stanca, A.M.; Morcia, C. Reducing the incidence of cereal head infection and mycotoxins in small grain cereal species. J. Cereal Sci. 2014, 59, 284–293. [Google Scholar] [CrossRef]
- Bai, G.; Shaner, G. Management and resistance in wheat and barley to Fusarium head blight. Annu. Rev. Phytopathol. 2004, 42, 135–161. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, H.; Christensen, J. Factors affecting resistance of wheat to scab caused by Gibberella zeae. Phytopathology 1963, 53, 831–838. [Google Scholar]
- Mesterhazy, A. Role of deoxynivalenol in aggressiveness of Fusarium graminearum and F. culmorum and in resistance to Fusarium head blight. Eur. J. Plant Pathol. 2002, 108, 675–684. [Google Scholar] [CrossRef]
- Miller, J.D.; Young, J.C.; Sampson, D.R. Deoxynivalenol and Fusarium head blight resistance in spring cereals. J. Phytopathol. 1985, 113, 359–367. [Google Scholar] [CrossRef]
- Buerstmayr, H.; Ban, T.; Anderson, J.A. QTL mapping and marker-assisted selection for Fusarium head blight resistance in wheat: A review. Plant Breed. 2009, 128, 1–26. [Google Scholar] [CrossRef]
- De la Pena, R.C.; Smith, K.P.; Capettini, F.; Muehlbauer, G.J.; Gallo-Meagher, M.; Dill-Macky, R.; Somers, D.A.; Rasmusson, D.C. Quantitative trait loci associated with resistance to Fusarium head blight and kernel discoloration in barley. Theor. Appl. Genet. 1999, 99, 561–569. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.Q.; Steffenson, B.J.; Prom, L.K.; Lapitan, N.L.V. Mapping of quantitative trait loci for Fusarium head blight resistance in barley. Phytopathology 2000, 90, 1079–1088. [Google Scholar] [CrossRef] [PubMed]
- Mesfin, A.; Smith, K.P.; Dill-Macky, R.; Evans, C.K.; Waugh, R.; Gustus, C.D.; Muehlbauer, G.J. Quantitative trait loci for Fusarium head blight resistance in barley detected in a two-rowed by six-rowed population. Crop Sci. 2003, 43, 307–318. [Google Scholar] [CrossRef]
- Martin, M.; Miedaner, T.; Dhillon, B.S.; Ufermann, U.; Kessel, B.; Ouzunova, M.; Schipprack, W.; Melchinger, A.E. Colocalization of qtl for gibberella ear rot resistance and low mycotoxin contamination in early European maize. Crop Sci. 2011, 51, 1935–1945. [Google Scholar] [CrossRef]
- Karlovsky, P. Biological detoxification of the mycotoxin deoxynivalenol and its use in genetically engineered crops and feed additives. Appl. Microbiol. Biotechnol. 2011, 91, 491–504. [Google Scholar] [CrossRef] [PubMed]
- Kluger, B.; Bueschl, C.; Lemmens, M.; Michlmayr, H.; Malachova, A.; Koutnik, A.; Maloku, I.; Berthiller, F.; Adam, G.; Krska, R.; et al. Biotransformation of the mycotoxin deoxynivalenol in Fusarium resistant and susceptible near isogenic wheat lines. PLoS ONE 2015. [Google Scholar] [CrossRef] [PubMed]
- Balmer, D.; Flors, V.; Glauser, G.; Mauch-Mani, B. Metabolomics of cereals under biotic stress: Current knowledge and techniques. Front. Plant Sci. 2013, 4, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Bollina, V.; Kumaraswamy, G.K.; Kushalappa, A.C.; Choo, T.M.; Dion, Y.; Rioux, S.; Faubert, D.; Hamzehzarghani, H. Mass spectrometry-based metabolomics application to identify quantitative resistance-related metabolites in barley against Fusarium head blight. Mol. Plant Pathol. 2010, 11, 769–781. [Google Scholar] [CrossRef] [PubMed]
- Bollina, V.; Kushalappa, A.C.; Choo, T.M.; Dion, Y.; Rioux, S. Identification of metabolites related to mechanisms of resistance in barley against Fusarium graminearum, based on mass spectrometry. Plant Mol. Biol. 2011, 77, 355–370. [Google Scholar] [CrossRef] [PubMed]
- Browne, R.A.; Brindle, K.M. H NMR-based metabolite profiling as a potential selection tool for breeding passive resistance against Fusarium head blight (FHB) in wheat. Mol. Plant Pathol. 2007, 8, 401–410. [Google Scholar] [CrossRef] [PubMed]
- Cajka, T.; Vaclavikova, M.; Dzuman, Z.; Vaclavik, L.; Ovesna, J.; Hajslova, J. Rapid metabolomics method based on liquid chromatography with mass spectrometry to study the Fusarium infection of barley. J. Sep. Sci. 2014, 37, 912–919. [Google Scholar] [CrossRef] [PubMed]
- Chamarthi, S.K.; Kumar, K.; Gunnaiah, R.; Kushalappa, A.C.; Dion, Y.; Choo, T.M. Identification of Fusarium head blight resistance related metabolites specific to doubled-haploid lines in barley. Eur. J. Plant Pathol. 2014, 138, 67–78. [Google Scholar] [CrossRef]
- Gunnaiah, R.; Kushalappa, A.C. Metabolomics deciphers the host resistance mechanisms in wheat cultivar Sumai-3, against trichothecene producing and non-producing isolates of Fusarium graminearum. Plant Physiol. Biochem. 2014, 83, 40–50. [Google Scholar] [CrossRef] [PubMed]
- Gunnaiah, R.; Kushalappa, A.C.; Duggavathi, R.; Fox, S.; Somers, D.J. Integrated metabolo-proteomic approach to decipher the mechanisms by which wheat QTL (FHB1) contributes to resistance against Fusarium graminearum. PLoS ONE 2012. [Google Scholar] [CrossRef] [PubMed]
- Hamzehzarghani, H.; Kushalappa, A.C.; Dion, Y.; Rioux, S.; Comeau, A.; Yaylayan, V.; Marshall, W.D.; Mather, D.E. Metabolic profiling and factor analysis to discriminate quantitative resistance in wheat cultivars against Fusarium head blight. Physiol. Mol. Plant Pathol. 2005, 66, 119–133. [Google Scholar] [CrossRef]
- Hamzehzarghani, H.; Paranidharan, V.; Abu-Nada, Y.; Kushalappa, A.C.; Dion, Y.; Rioux, S.; Comeau, A.; Yaylayan, V.; Marshall, W.D. Metabolite profiling coupled with statistical analyses for potential high-throughput screening of quantitative resistance to Fusarium head blight in wheat. Can. J. Plant Pathol. 2008, 30, 24–36. [Google Scholar] [CrossRef]
- Kumaraswamy, G.K. Differential metabolic response of barley genotypes, varying in resistance, to trichothecene-producing and -nonproducing (tri5-) isolates of Fusarium graminearum. Plant Pathol. 2012, 61, 509–521. [Google Scholar] [CrossRef]
- Kumaraswamy, G.K.; Bollina, V.; Kushalappa, A.C.; Choo, T.M.; Dion, Y.; Rioux, S.; Mamer, O.; Faubert, D. Metabolomics technology to phenotype resistance in barley against gibberella zeae. Eur. J. Plant Pathol. 2011, 130, 29–43. [Google Scholar] [CrossRef]
- Kumaraswamy, K.G.; Kushalappa, A.C.; Choo, T.M.; Dion, Y.; Rioux, S. Mass spectrometry based metabolomics to identify potential biomarkers for resistance in barley against Fusarium head blight (Fusarium graminearum). J. Chem. Ecol. 2011, 37, 846–856. [Google Scholar] [CrossRef] [PubMed]
- Paranidharan, V.; Abu-Nada, Y.; Hamzehzarghani, H.; Kushalappa, A.C.; Mamer, O.; Dion, Y.; Rioux, S.; Comeau, A.; Choiniere, L. Resistance-related metabolites in wheat against Fusarium graminearum and the virulence factor deoxynivalenol (DON). Botany 2008, 86, 1168–1179. [Google Scholar] [CrossRef]
- Heuberger, A.L.; Robison, F.M.; Lyons, S.M.; Broeckling, C.D.; Prenni, J.E. Evaluating plant immunity using mass spectrometry-based metabolomics workflows. Front. Plant Sci. 2014, 5, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Bino, R.J.; Hall, R.D.; Fiehn, O.; Kopka, J.; Saito, K.; Draper, J.; Nikolau, B.J.; Mendes, P.; Roessner-Tunali, U.; Beale, M.H.; et al. Potential of metabolomics as a functional genomics tool. Trends Plant Sci. 2004, 9, 418–425. [Google Scholar] [CrossRef] [PubMed]
- Fiehn, O.; Kopka, J.; Dörmann, P.; Altmann, T.; Trethewey, R.N.; Willmitzer1, L. Metabolite profiling for plant functional genomics. Nat. Biotechnol. 2000, 18, 1157–1161. [Google Scholar] [CrossRef] [PubMed]
- Hall, R.; Beale, M.; Fiehn, O.; Hardy, N.; Sumner, L.; Bino, R. Plant metabolomics: The missing link in functional genomics strategies. Plant Cell 2002, 14, 1437–1440. [Google Scholar] [CrossRef] [PubMed]
- Sumner, L.W.; Mendes, P.; Dixon, R.A. Plant metabolomics: Large-scale phytochemistry in the functional genomics era. Phytochemistry 2003, 62, 817–836. [Google Scholar] [CrossRef]
- Castro, C.; Manetti, C. A multiway approach to analyze metabonomic data: A study of maize seeds development. Anal. Biochem. 2007, 371, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, J.K.; Connelly, J.; Lindon, J.C.; Holmes, E. Metabonomics: A platform for studying drug toxicity and gene function. Nat. Rev. Drug Discov. 2002, 1, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Ward, J.L.; Harris, C.; Lewis, J.; Beale, M.H. Assessment of 1h nmr spectroscopy and multivariate analysis as a technique for metabolite fingerprinting of Arabidopsis thaliana. Phytochemistry 2003, 62, 949–957. [Google Scholar] [CrossRef]
- Warth, B.; Parich, A.; Bueschl, C.; Schoefbeck, D.; Neumann, N.K.; Kluger, B.; Schuster, K.; Krska, R.; Adam, G.; Lemmens, M.; et al. GC-MS based targeted metabolic profiling identifies changes in the wheat metabolome following deoxynivalenol treatment. Metabolomics 2015, 11, 722–738. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.; Veyrat, N.; Gordon-Weeks, R.; Zhang, Y.H.; Martin, J.; Smart, L.; Glauser, G.; Erb, M.; Flors, V.; Frey, M.; et al. Benzoxazinoid metabolites regulate innate immunity against aphids and fungi in maize. Plant Physiol. 2011, 157, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Niemeyer, H.M. Hydroxamic acids derived from 2-hydroxy-2H-1,4-benzoxazin-3(4H)-one: Key defense chemicals of cereals. J. Agric. Food Chem. 2009, 57, 1677–1696. [Google Scholar] [CrossRef] [PubMed]
- Zadoks, J.C.; Chang, T.T.; Konzac, C.F. A decimal code for the growth stages of cereals. Weed Res. 1974, 14, 415–421. [Google Scholar] [CrossRef]
- Lattanzio, F. Role of phenolics in the resistance mechanisms of plants against fungal pathogens and insects. In Phytochemistry: Advances in Research; Research Signpost: Trivandrum, Kerala, India, 2006; pp. 23–67. [Google Scholar]
- Dixon, R.A.; Achnine, L.; Kota, P.; Liu, C.-J.; Reddy, M.S.S.; Wang, L. The phenylpropanoid pathway and plant defence—A genomics perspective. Mol. Plant Pathol. 2002, 3, 371–390. [Google Scholar] [CrossRef] [PubMed]
- Ravensdale, M.; Rocheleau, H.; Wang, L.; Nasmith, C.; Ouellet, T.; Subramaniam, R. Components of priming-induced resistance to Fusarium head blight in wheat revealed by two distinct mutants of Fusarium graminearum. Mol. Plant Pathol. 2014, 15, 948–956. [Google Scholar] [CrossRef] [PubMed]
- Buśko, M.; Góral, T.; Ostrowska, A.; Matysiak, A.; Walentyn-Góral, D.; Perkowski, J. The effect of Fusarium inoculation and fungicide application on concentrations of flavonoids (apigenin, kaempferol, luteolin, naringenin, quercetin, rutin, vitexin) in winter wheat cultivars. Am. J. Plant Sci. 2014, 5, 3727–3736. [Google Scholar] [CrossRef]
- Agati, G.; Azzarello, E.; Pollastri, S.; Tattini, M. Flavonoids as antioxidants in plants: Location and functional significance. Plant Sci. 2012, 196, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, I.; Alegre, L.; van Breusegem, F.; Munne-Bosch, S. How relevant are flavonoids as antioxidants in plants? Trends Plant Sci. 2009, 14, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Jung, M.J.; Heo, S.-I.; Wang, M.-H. HPLC analysis and antioxidant activity of Ulmus davidiana and some flavonoids. Food Chem. 2010, 120, 313–318. [Google Scholar] [CrossRef]
- Treutter, D. Significance of flavonoids in plant resistance: A review. Environ. Chem. Lett. 2006, 4, 147–157. [Google Scholar] [CrossRef]
- Venturini, G.; Toffolatti, S.L.; Assante, G.; Babazadeh, L.; Campia, P.; Fasoli, E.; Salomoni, D.; Vercesi, A. The influence of flavonoids in maize pericarp on Fusarium ear rot symptoms and fumonisin accumulation under field conditions. Plant Pathol. 2015, 64, 671–679. [Google Scholar] [CrossRef]
- Treutter, D. Significance of flavonoids in plant resistance and enhancement of their biosynthesis. Plant Biol. 2005, 7, 581–591. [Google Scholar] [CrossRef] [PubMed]
- Mierziak, J.; Kostyn, K.; Kulma, A. Flavonoids as important molecules of plant interactions with the environment. Molecules 2014, 19, 16240–16265. [Google Scholar] [CrossRef] [PubMed]
- Eggert, K.; Hollmann, J.; Hiller, B.; Kruse, H.P.; Rawel, H.M.; Pawelzik, E. Effects of Fusarium infection on the phenolics in emmer and naked barley. J. Agric. Food Chem. 2010, 58, 3043–3049. [Google Scholar] [CrossRef] [PubMed]
- Chitarrini, G.; Nobili, C.; Pinzari, F.; Antonini, A.; de Rossi, P.; del Fiore, A.; Procacci, S.; Tolaini, V.; Scala, V.; Scarpari, M.; et al. Buckwheat achenes antioxidant profile modulates Aspergillus flavu growth and aflatoxin production. Int. J. Food Microbiol. 2014, 189, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Norton, R.A. Inhibition of aflatoxin B1 biosynthesis in Aspergillus flavu by anthocyanidins and related flavonoids. J. Agric. Food Chem. 1999, 47, 1230–1235. [Google Scholar] [CrossRef] [PubMed]
- Salas, M.P.; Reynoso, C.M.; Céliz, G.; Daz, M.; Resnik, S.L. Efficacy of flavanones obtained from citrus residues to prevent patulin contamination. Food Res. Int. 2012, 48, 930–934. [Google Scholar] [CrossRef]
- Desjardins, A.E.; Plattner, R.D.; Spencer, G.F. Inhibition of trichothecene toxin biosynthesis by naturally-occurring shikimate aromatics. Phytochemistry 1988, 27, 767–771. [Google Scholar] [CrossRef]
- Fernandez-Orozco, R.; Li, L.; Harflett, C.; Shewry, P.R.; Ward, J.L. Effects of environment and genotype on phenolic acids in wheat in the healthgrain diversity screen. J. Agric. Food Chem. 2010, 58, 9341–9352. [Google Scholar] [CrossRef] [PubMed]
- Assabgui, R.A.; Reid, L.M.; Hamilton, R.I.; Arnason, J.T. Correlation of kernel (E)-ferulic acid content maize with resistance to Fusarium graminearum. Phytopathology 1993, 83, 949–953. [Google Scholar] [CrossRef]
- Bily, A.C.; Reid, L.M.; Taylor, J.H. Dehydrodimers of ferulic acid in maize grain pericarp and aleurone: Resistance factors to Fusarium graminearum. Phytopathology 2003, 93, 712–719. [Google Scholar] [CrossRef] [PubMed]
- Samapundo, S.; de Meulenaer, B.; Osei-Nimoh, D.; Lamboni, Y.; Debevere, J.; Devlieghere, F. Can phenolic compounds be used for the protection of corn from fungal invasion and mycotoxin contamination during storage? Food Microbiol. 2007, 24, 465–473. [Google Scholar] [CrossRef] [PubMed]
- McKeehen, J.D.; Busch, R.H.; Fulcher, R.G. Evaluation of wheat (Triticum aestivum L.) phenolic acids during grain development and their contribution to Fusarium resistance. J. Agric. Food Chem. 1999, 47, 1476–1482. [Google Scholar] [CrossRef] [PubMed]
- Siranidou, E.; Kang, Z.; Buchenauer, H. Studies on symptom development, phenolic compounds and morphological defence responses in wheat cultivars differing in resistance to Fusarium head blight. J. Phytopathol. 2002, 150, 200–208. [Google Scholar] [CrossRef]
- Nicholson, R.L.; Hammerschmidt, R. Phenolic compounds and their role in disease resistance. Annu. Rev. Phytopathol. 1992, 30, 369–389. [Google Scholar] [CrossRef]
- Boutigny, A.-L.; Richard-Forget, F.; Barreau, C. Natural mechanisms for cereal resistance to the accumulation of Fusarium trichothecenes. Eur. J. Plant Pathol. 2008, 121, 411–423. [Google Scholar] [CrossRef]
- Boutigny, A.L.; Barreau, C.; Atanasova-Penichon, V.; Verdal-Bonnin, M.N.; Pinson-Gadais, L.; Richard-Forget, F. Ferulic acid, an efficient inhibitor of type B trichothecene biosynthesis and Tri gene expression in Fusarium liquid cultures. Mycol. Res. 2009, 113, 746–753. [Google Scholar] [CrossRef] [PubMed]
- Ponts, N.; Pinson-Gadais, L.; Boutigny, A.L.; Barreau, C.; Richard-Forget, F. Cinnamic-derived acids significantly affect Fusarium graminearum growth and in vitro synthesis of type B trichothecenes. Phytopathology 2011, 101, 929–934. [Google Scholar] [CrossRef] [PubMed]
- Bakan, B.; Bily, A.; Melcion, D.; Cahagnier, B.; Regnault-Roger, C.; Philogene, B.; Richard-Molard, D. Possible role of plant phenolics in the production of trichothecenes by Fusarium graminearum strains on different fractions of maize kernels. J. Agric. Food Chem. 2003, 51, 2826–2831. [Google Scholar] [CrossRef] [PubMed]
- Beekrum, S.; Govinden, R.; Padayachee, T.; Odhav, B. Naturally occurring phenols: A detoxification strategy for fumonisin B1. Food. Addit. Contam. 2003, 20, 490–493. [Google Scholar] [PubMed]
- Kostyn, K.; Czemplik, M.; Kulma, A.; Bortniczuk, M.; Skala, J.; Szopa, J. Genes of phenylpropanoid pathway are activated in early response to Fusarium attack in flax plants. Plant Sci. 2012, 190, 103–115. [Google Scholar] [CrossRef] [PubMed]
- Boutigny, A.-L. Analyse de facteurs biochimiques interagissant dans le processus de biosynthèse des TCTB. In Proceedings of Colloque Fusariotoxines des Céréale, Arcachon, France, 11–13 September 2007.
- Ponts, N.; Pinson-Gadais, L.; Barreau, C.; Richard-Forget, F.; Ouellet, T. Exogenous H2O2 and catalase treatments interfere with tri genes expression in liquid cultures of Fusarium graminearum. FEBS Lett. 2007, 581, 443–447. [Google Scholar] [CrossRef] [PubMed]
- Passone, M.A.; Ruffino, M.; Ponzio, V.; Resnik, S.; Etcheverry, M.G. Postharvest control of peanut Aspergillus section Flavi populations by a formulation of food-grade antioxidants. Int. J. Food Microbiol. 2009, 131, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Lionetti, V.; Giancaspro, A.; Fabri, E.; Giove, S.L.; Reem, N.; Zabotina, O.A.; Blanco, A.; Gadaleta, A.; Bellincampi, D. Cell wall traits as potential resources to improve resistance of durum wheat against Fusarium graminearum. BMC Plant Biol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Naoumkina, M.A.; Zhao, Q.; Gallego-Giraldo, L.; Dai, X.; Zhao, P.X.; Dixon, R.A. Genome-wide analysis of phenylpropanoid defence pathways. Mol. Plant Pathol. 2010, 11, 829–846. [Google Scholar] [CrossRef] [PubMed]
- Sattler, S.E.; Funnell-Harris, D.L. Modifying lignin to improve bioenergy feedstocks: Strengthening the barrier against pathogens? Front. Plant Sci. 2013, 4, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.Y.; Choi, G.J.; Son, S.W.; Jang, K.S.; Lim, H.K.; Lee, S.O.; Sung, N.D.; Cho, K.Y.; Kim, J.-C. Isolation and antifungal activity of lignans from Myristica fragrans against various plant pathogenic fungi. Pest Manag. Sci. 2007, 63, 935–940. [Google Scholar] [CrossRef] [PubMed]
- Kachroo, A.; Kachroo, P. Fatty acid-derived signals in plant defense. Annu. Rev. Phytopathol. 2009, 47, 153–176. [Google Scholar] [CrossRef] [PubMed]
- Ongena, M.; Duby, F.; Rossignol, F.; Fauconnier, M.-L.; Dommes, J.; Thonart, P. Stimulation of the lipoxygenase pathway is associated with systemic resistance induced in bean by a nonpathogenic Pseudomonas strain. Mol. Plant Microbe Interact. 2004, 17, 1009–1018. [Google Scholar] [CrossRef] [PubMed]
- Walters, D.; Raynor, L.; Mitchell, A.; Walker, R.; Walker, K. Antifungal activities of four fatty acids against plant pathogenic fungi. Mycopathologia 2004, 157, 87–90. [Google Scholar] [PubMed]
- Burow, G.B.; Nesbitt, T.C.; Dunlap, J.; Keller, N.P. Seed lipoxygenase products modulate Aspergillus mycotoxin biosynthesis. Mol. Plant Microbe Interact. 1997, 10, 380–387. [Google Scholar] [CrossRef]
- Tiwari, R.P.; Mittal, V.; Singh, G.; Bhalla, T.C.; Saini, S.S.; Vadehra, D.V. Effect of fatty-acids on aflatoxin production by Aspergillus parasiticus. Folia Microbiol. 1986, 31, 120–123. [Google Scholar] [CrossRef]
- Yaeno, T.; Matsuda, O.; Iba, K. Role of chloroplast trienoic fatty acids in plant disease defense responses. Plant J. 2004, 40, 931–941. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.Q.; Shim, W.B.; Gobel, C.; Kunze, S.; Feussner, I.; Meeley, R.; Balint-Kurti, P.; Kolomiets, M. Disruption of a maize 9-lipoxygenase results in increased resistance to fungal pathogens and reduced levels of contamination with mycotoxin fumonisin. Mol. Plant Microbe Interact. 2007, 20, 922–933. [Google Scholar] [CrossRef] [PubMed]
- Morcillo, R.J.L.; Ocampo, J.A.; Garrido, J.M.G. Plant 9-lox oxylipin metabolism in response to arbuscular mycorrhiza. Plant Signal. Behav. 2012, 7, 1584–1588. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Brodhagen, M.; Isakeit, T.; Brown, S.H.; Göbel, C.; Betran, J.; Feussner, I.; Keller, N.P.; Kolomiets, M.V. Inactivation of the lipoxygenase ZmLOX3 increases susceptibility of maize to Aspergillus spp. Mol. Plant Microbe Interact. 2009, 22, 222–231. [Google Scholar] [CrossRef] [PubMed]
- Fabbri, A.A.; Fanelli, C.; Panfili, G.; Passi, S.; Fasella, P. Lipoperoxidation and aflatoxin biosynthesis by Aspergillus parasiticus and A. Flavus. J. Gen. Microbiol. 1983, 129, 3447–3452. [Google Scholar] [CrossRef]
- Fanelli, C.; Fabbri, A.A. Relationship between lipids and aflatoxin biosynthesis. Mycopathologia 1989, 107, 115–120. [Google Scholar] [CrossRef] [PubMed]
- Passi, S.; Nazzaroporro, M.; Fanelli, C.; Fabbri, A.A.; Fasella, P. Role of lipoperoxidation in aflatoxin production. Appl. Microbiol. Biotechnol. 1984, 19, 186–190. [Google Scholar] [CrossRef]
- Brodhagen, M.; Keller, N.P. Signalling pathways connecting mycotoxin production and sporulation. Mol. Plant Pathol. 2006, 7, 285–301. [Google Scholar] [CrossRef] [PubMed]
- Nobili, C.; D’Angeli, S.; Altamura, M.M.; Scala, V.; Fabbri, A.A.; Reverberi, M.; Fanelli, C. ROS and 9-oxylipins are correlated with deoxynivalenol accumulation in the germinating caryopses of Triticum aestivum after Fusarium graminearum infection. Eur. J. Plant Pathol. 2014, 139, 429–444. [Google Scholar] [CrossRef]
- Li, G.; Yen, Y. Jasmonate and ethylene signaling pathway may mediate Fusarium head blight resistance in wheat. Crop Sci. 2008, 48, 1888–1996. [Google Scholar] [CrossRef]
- Zhang, L.; Xing, D. Methyl jasmonate induces production of reactive oxygen species and alterations in mitochondrial dynamics that precede photosynthetic dysfunction and subsequent cell death. Plant Cell Physiol. 2008, 49, 1092–1110. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K.; Manners, J.M. Jasmonate signaling: Toward an integrated view. Plant Physiol. 2008, 146, 1459–1468. [Google Scholar] [CrossRef] [PubMed]
- Bari, R.; Jones, J.D. Role of plant hormones in plant defence responses. Plant Mol. Biol. 2009, 69, 473–488. [Google Scholar] [CrossRef] [PubMed]
- Glazebrook, J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 2005, 43, 205–227. [Google Scholar] [CrossRef] [PubMed]
- Makandar, R.; Nalam, V.J.; Lee, H.; Trick, H.N.; Dong, Y.; Shah, J. Salicylic acid regulates basal resistance to Fusarium head blight in wheat. Mol. Plant Microbe Interact. 2012, 25, 431–439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poppenberger, B.; Berthiller, F.; Lucyshyn, D.; Sieberer, T.; Schuhmacher, R.; Krska, R.; Kuchler, K.; Glossl, J.; Luschnig, C.; Adam, G. Detoxification of the Fusarium mycotoxin deoxynivalenol by a UDP-glucosyltransferase from Arabidopsis thaliana. J. Biol. Chem. 2003, 278, 47905–47914. [Google Scholar] [CrossRef] [PubMed]
- Goodrich-Tanrikulu, M.; Mahoney, N.E.; Rodriguez, S.B. The plant growth regulator methyl jasmonate inhibits aflatoxin production by Aspergillus flavu. Microbiology 1995, 141, 2831–2837. [Google Scholar] [CrossRef] [PubMed]
- Meimaroglou, D.M.; Galanopoulou, D.; Flouri, F.; Panagiota, M. The plant growth regulator methyl jasmonate inhibits aflatoxin B1 production by Aspergillus parasiticus in caper. Int. J. Nutr. Food Sci. 2015, 10–17. [Google Scholar]
- Vergopoulou, S.; Galanopoulou, D.; Markaki, P. Methyl jasmonate stimulates aflatoxin B1 biosynthesis by Aspergillus parasiticus. J. Agric. Food Chem. 2001, 49, 3494–3498. [Google Scholar] [CrossRef] [PubMed]
- Petti, C.; Reiber, K.; Ali, S.S.; Berney, M.; Doohan, F.M. Auxin as a player in the biocontrol of Fusarium head blight disease of barley and its potential as a disease control agent. BMC Plant Biol. 2012, 12, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Mauch-Mani, B.; Mauch, F. The role of abscisic acid in plant-pathogen interactions. Curr. Opin. Plant Biol. 2005, 8, 409–414. [Google Scholar] [CrossRef] [PubMed]
- Flors, V.; Ton, J.; Jakab, G.; Mauch-Mani, B. Abscisic acid and callose: Team players in defence against pathogens? J. Phytopathol. 2005, 153, 377–383. [Google Scholar] [CrossRef]
- Kang, Z.S.; Buchenauer, H. Cytology and ultrastructure of the infection of wheat spikes by Fusarium culmorum. Mycol. Res. 2000, 104, 1083–1093. [Google Scholar] [CrossRef]
- Chen, X.; Steed, A.; Travella, S.; Keller, B.; Nicholson, P. Fusarium graminearum exploits ethylene signalling to colonize dicotyledonous and monocotyledonous plants. New Phytol. 2009, 182, 975–983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmelz, E.A.; Huffaker, A.; Sims, J.W.; Christensen, S.A.; Lu, X.; Okada, K.; Peters, R.J. Biosynthesis, elicitation and roles of monocot terpenoid phytoalexins. Plant J. 2014, 79, 659–678. [Google Scholar] [CrossRef] [PubMed]
- Piesik, D.; Panka, D.; Delaney, K.J.; Skoczek, A.; Lamparski, R.; Weaver, D.K. Cereal crop volatile organic compound induction after mechanical injury, beetle herbivory (Oulema spp.), or fungal infection (Fusarium spp.). J. Plant Physiol. 2011, 168, 878–886. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, I.T.; Halitschke, R.; Paschold, A.; von Dahl, C.C.; Preston, C.A. Volatile signaling in plant-plant interactions: “Talking trees” in the genomics era. Science 2006, 311, 812–815. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Eudes, F.; Laroche, A. Identification of differentially regulated proteins in response to a compatible interaction between the pathogen Fusarium graminearum and its host, Triticum aestivum. Proteomics 2006, 6, 4599–4609. [Google Scholar] [CrossRef] [PubMed]
- Tzin, V.; Galili, G. New insights into the shikimate and aromatic amino acids biosynthesis pathways in plants. Mol. Plant 2010, 3, 956–972. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, N.J.; Lilley, C.J.; Urwin, P.E. Identification of genes involved in the response of Arabidopsis to simultaneous biotic and abiotic stesses. Plant Physiol. 2013, 162, 2028–2041. [Google Scholar] [CrossRef] [PubMed]
- Montesano, M.; Brader, G.; Palva, E.T. Pathogen derived elicitors: Searching for receptors in plants. Mol. Plant Pathol. 2003, 4, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Walters, D. Resistance to plant pathogens: Possible roles for free polyamines and polyamine catabolism. New Phytol. 2003, 159, 109–115. [Google Scholar] [CrossRef]
- Campos-Bermudez, V.A.; Fauguel, C.M.; Tronconi, M.A.; Casati, P.; Presello, D.A.; Andreo, C.S. Transcriptional and metabolic changes associated to the infection by Fusarium verticillioides in maize inbreds with contrasting Ear Rot resistance. PLoS ONE 2013, 4, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Wojtasik, W.; Kulma, A.; Namysl, K.; Preisner, M.; Szopa, J. Polyamine metabolism in flax in response to treatment with pathogenic and non-pathogenic Fusarium strains. Front. Plant Sci. 2015, 6, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Bremont, J.F.; Marina, M.; Guerrero-Gonzalez, M.D.; Rossi, F.R.; Sanchez-Rangel, D.; Rodriguez-Kessler, M.; Ruiz, O.; Garriz, A. Physiological and molecular implications of plant polyamine metabolism during biotic interactions. Front. Plant Sci. 2014, 5, 1–14. [Google Scholar]
- Stoessl, A. The antifungal factors in barley. IV. Isolation, structure and synthesis of the hordatines. Can. J. Chem. 1967, 45, 1745–1760. [Google Scholar] [CrossRef]
- Gardiner, D.M.; Kazan, K.; Praud, S.; Torney, F.J.; Rusu, A.; Manners, J.M. Early activation of wheat polyamine biosynthesis during Fusarium head blight implicates putrescine as an inducer of trichothecene mycotoxin production. BMC Plant Biol. 2010, 10, 1471–2229. [Google Scholar] [CrossRef] [PubMed]
- Crespo-Sempere, A.; Estiarte, N.; Marin, S.; Sanchis, V.; Ramos, A.J. Targeting Fusarium graminearum control via polyamine enzyme inhibitors and polyamine analogs. Food Microbiol. 2015, 49, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Gillaspy, G.E. The cellular language of myo-inositol signaling. New Phytol. 2011, 192, 823–839. [Google Scholar] [CrossRef] [PubMed]
- Loewus, F.A. Inositol and Plant Cell Wall Polysaccharide Biogenesis. In Subcellular Biochemistry; Majumder, A.L., Biswas, B.B., Eds.; Springer: Pullman, WA, USA, 2006; Volume 39, pp. 21–45. [Google Scholar]
- Lorence, A.; Chevone, B.I.; Mendes, P.; Nessler, C.L. myo-Inositol oxygenase offers a possible entry point into plant ascorbate biosynthesis. Plant Physiol. 2004, 134, 1200–1205. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.Y.; Ding, L.L.; Li, J.M.; Xu, B.L.; Yang, L.; Bi, K.S.; Wang, Z.T. 1H NMR and MS based metabolomics study of the intervention effect of curcumin on hyperlipidemia mice induced by high-fat diet. PLoS ONE 2015. [Google Scholar] [CrossRef] [PubMed]
- Kushalappa, A.C.; Gunnaiah, R. Metabolo-proteomics to discover plant biotic stress resistance genes. Trends Plant Sci. 2013, 18, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Gartner, T.; Steinfath, M.; Andorf, S.; Lisec, J.; Meyer, R.C.; Altmann, T.; Willmitzer, L.; Selbig, J. Improved heterosis prediction by combining information on DNA- and metabolic markers. PLoS ONE 2009. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gauthier, L.; Atanasova-Penichon, V.; Chéreau, S.; Richard-Forget, F. Metabolomics to Decipher the Chemical Defense of Cereals against Fusarium graminearum and Deoxynivalenol Accumulation. Int. J. Mol. Sci. 2015, 16, 24839-24872. https://doi.org/10.3390/ijms161024839
Gauthier L, Atanasova-Penichon V, Chéreau S, Richard-Forget F. Metabolomics to Decipher the Chemical Defense of Cereals against Fusarium graminearum and Deoxynivalenol Accumulation. International Journal of Molecular Sciences. 2015; 16(10):24839-24872. https://doi.org/10.3390/ijms161024839
Chicago/Turabian StyleGauthier, Léa, Vessela Atanasova-Penichon, Sylvain Chéreau, and Florence Richard-Forget. 2015. "Metabolomics to Decipher the Chemical Defense of Cereals against Fusarium graminearum and Deoxynivalenol Accumulation" International Journal of Molecular Sciences 16, no. 10: 24839-24872. https://doi.org/10.3390/ijms161024839