A Large-Scale Analysis of the Relationship of Synonymous SNPs Changing MicroRNA Regulation with Functionality and Disease
Abstract
:1. Introduction
2. Results
2.1. The Putative Model for the Contributions of Synonymous Mutations to Functionality and Disease by miRNA-Mediated Gene Regulation
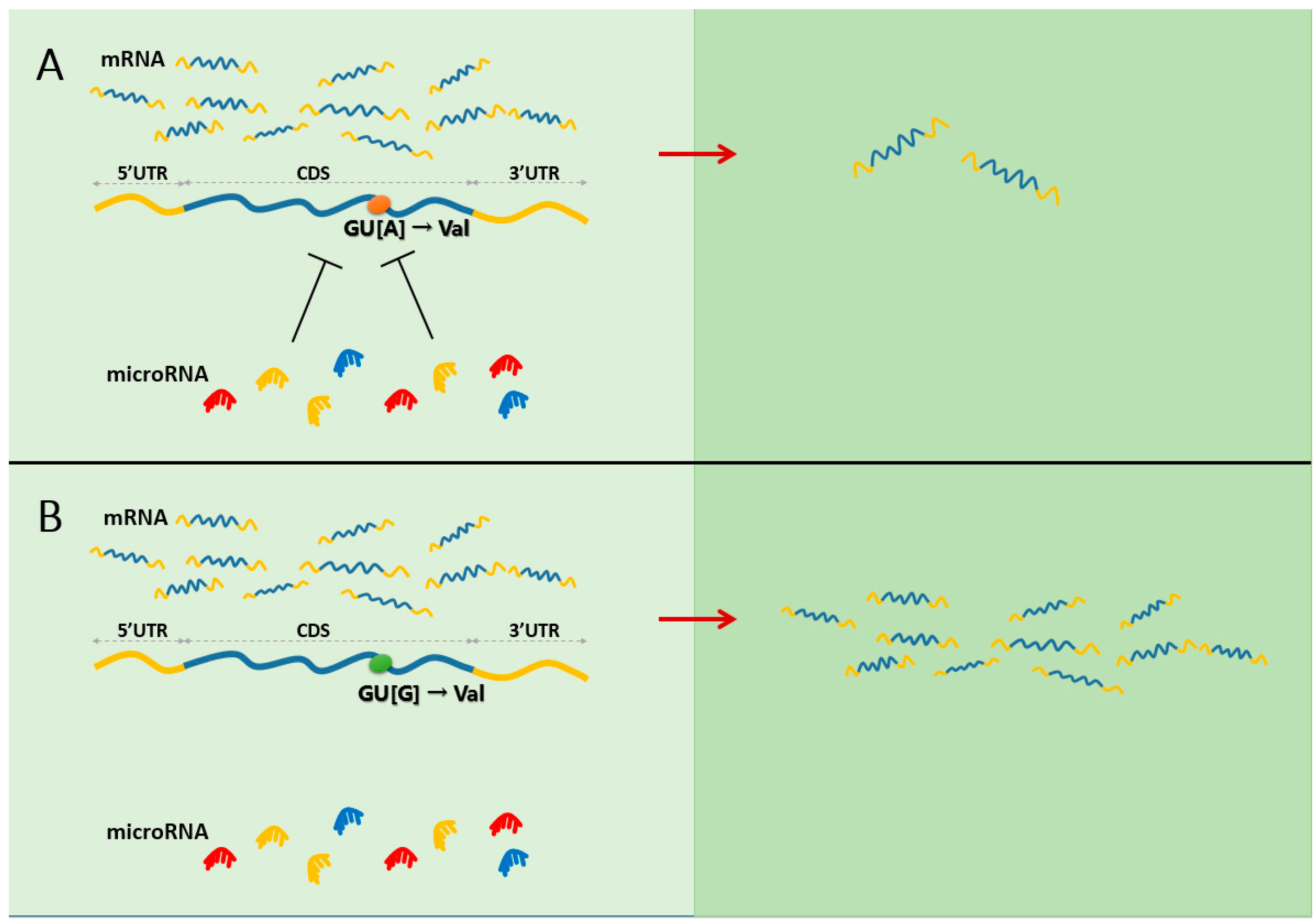
2.2. Experimental Evidence Supporting the Model
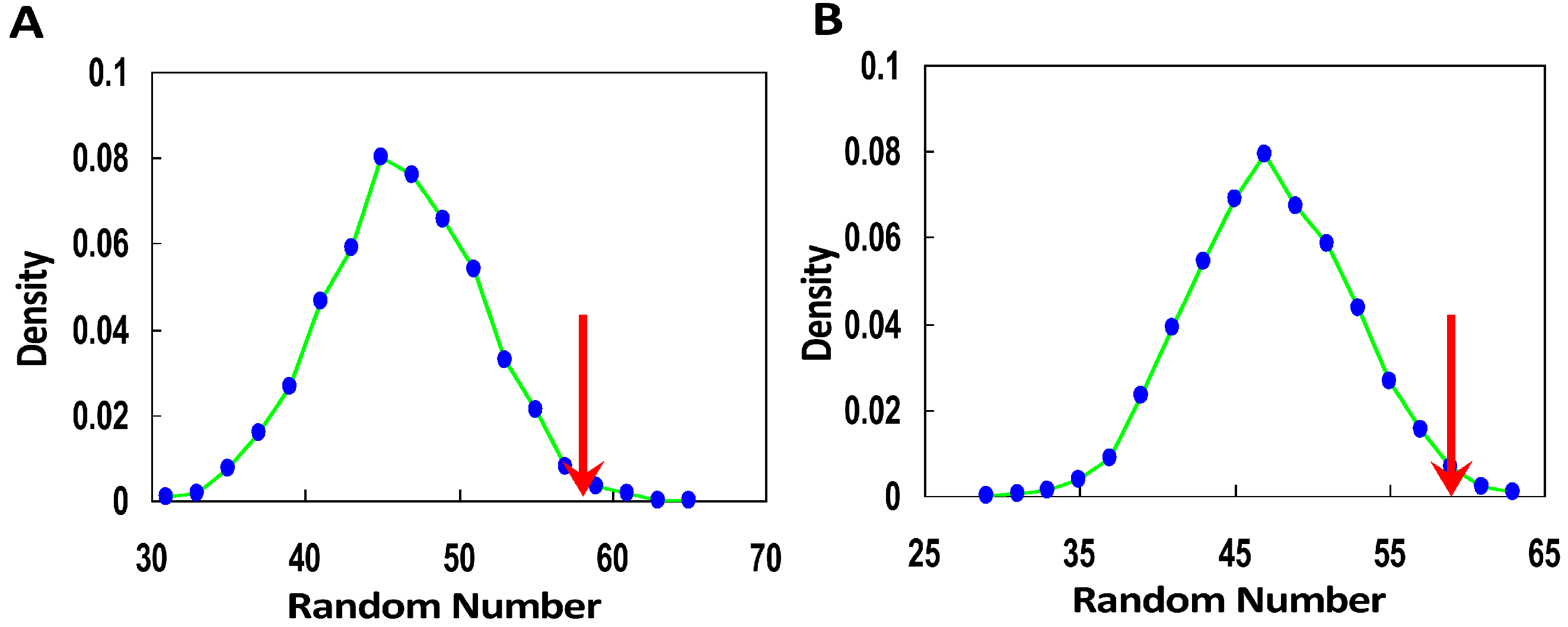
2.3. Selection Signal Evidence from the Integrated Haplotype Score to Support the Model
| Population | p-Value | Recent Selection | A | B |
|---|---|---|---|---|
| ASI | 4.7 × 10−4 | C | 58 | 44 |
| D | 3212 | 4972 | ||
| YRI | 0.02 | C | 79 | 63 |
| D | 4449 | 5259 | ||
| CEU | 0.46 | C | 51 | 71 |
| D | 3954 | 4765 | ||
| Genotype & Expression | 0.02 | E | 59 | 45 |
| F | 21,237 | 25,698 |
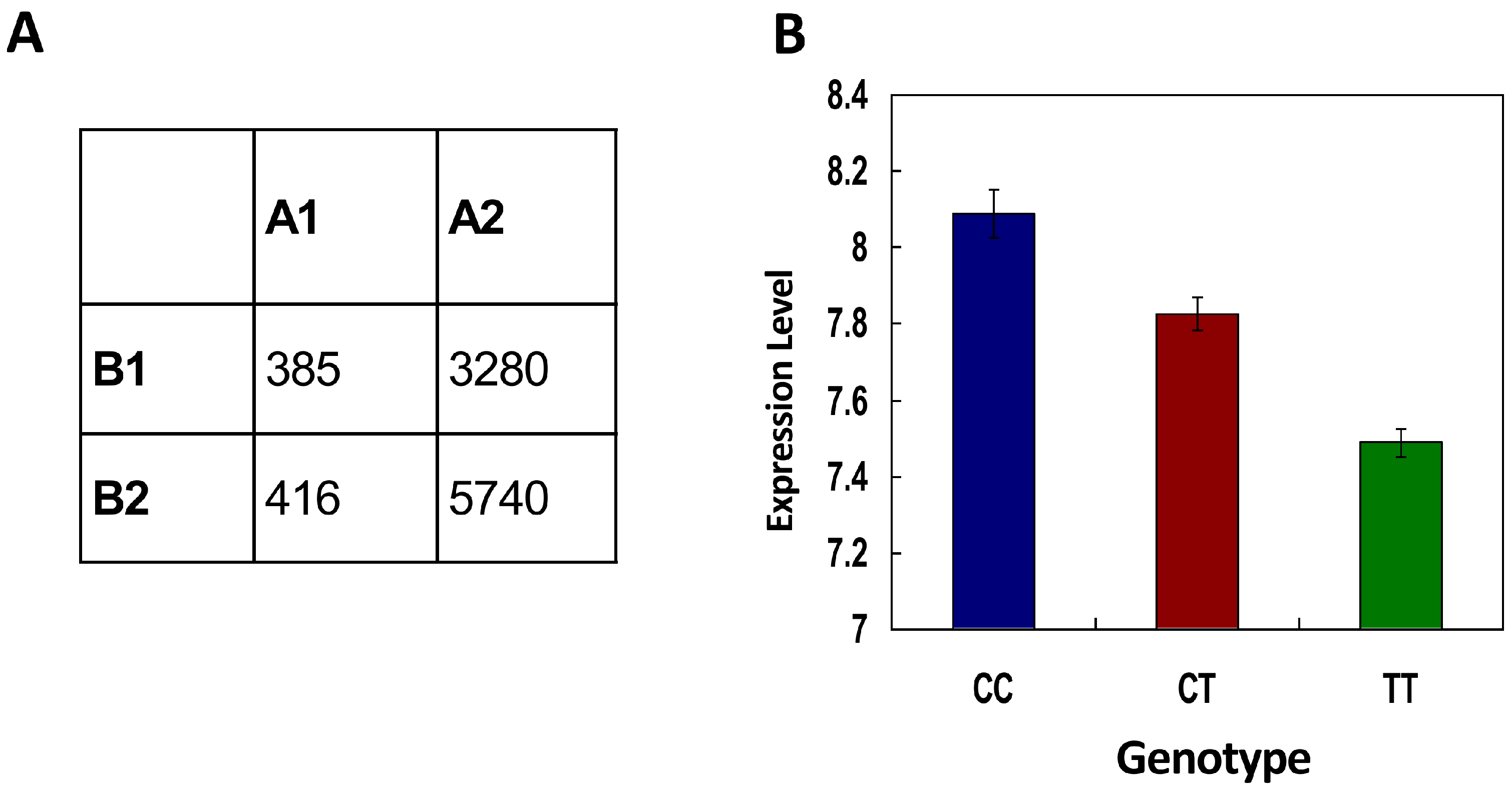
2.4. Gene Expression Evidence to Support the Model
2.5. Genome-Wide Association Loci Evidence for Human Disease and Trait to Support the Model
2.6. Extension of the Model
3. Discussion
4. Experimental Section
4.1. Data Used in this Study
4.2. Tools for Data Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Schonrock, N.; Harvey, R.P.; Mattick, J.S. Long noncoding RNAs in cardiac development and pathophysiology. Circ. Res. 2012, 111, 1349–1362. [Google Scholar] [PubMed]
- Sauna, Z.E.; Kimchi-Sarfaty, C. Understanding the contribution of synonymous mutations to human disease. Nat. Rev. Genet. 2011, 12, 683–691. [Google Scholar] [CrossRef] [PubMed]
- Chamary, J.V.; Parmley, J.L.; Hurst, L.D. Hearing silence: Non-neutral evolution at synonymous sites in mammals. Nat. Rev. Genet. 2006, 7, 98–108. [Google Scholar] [CrossRef] [PubMed]
- Pagani, F.; Raponi, M.; Baralle, F.E. Synonymous mutations in CFTR exon 12 affect splicing and are not neutral in evolution. Proc. Natl. Acad. Sci. USA 2005, 102, 6368–6372. [Google Scholar] [CrossRef] [PubMed]
- Kimchi-Sarfaty, C.; Oh, J.M.; Kim, I.W.; Sauna, Z.E.; Calcagno, A.M.; Ambudkar, S.V.; Gottesman, M.M. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science 2007, 315, 525–528. [Google Scholar] [CrossRef] [PubMed]
- Hindorff, L.A.; Sethupathy, P.; Junkins, H.A.; Ramos, E.M.; Mehta, J.P.; Collins, F.S.; Manolio, T.A. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. USA 2009, 106, 9362–9367. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Griffiths-Jones, S. The microRNA Registry. Nucleic Acids Res. 2004, 32, D109–D111. [Google Scholar] [CrossRef] [PubMed]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Zhang, Q.; Deng, M.; Miao, J.; Guo, Y.; Gao, W.; Cui, Q. An analysis of human microRNA and disease associations. PLoS ONE 2008, 3, e3420. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Croce, C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer 2006, 6, 857–866. [Google Scholar] [CrossRef] [PubMed]
- Latronico, M.V.; Catalucci, D.; Condorelli, G. Emerging role of microRNAs in cardiovascular biology. Circ. Res. 2007, 101, 1225–1236. [Google Scholar] [CrossRef] [PubMed]
- Helwak, A.; Kudla, G.; Dudnakova, T.; Tollervey, D. Mapping the Human miRNA Interactome by CLASH Reveals Frequent Noncanonical Binding. Cell 2013, 153, 654–665. [Google Scholar] [CrossRef] [PubMed]
- Grimson, A.; Farh, K.K.; Johnston, W.K.; Garrett-Engele, P.; Lim, L.P.; Bartel, D.P. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol. Cell 2007, 27, 91–105. [Google Scholar] [CrossRef] [PubMed]
- Grey, F.; Tirabassi, R.; Meyers, H.; Wu, G.; McWeeney, S.; Hook, L.; Nelson, J.A. A viral microRNA down-regulates multiple cell cycle genes through mRNA 5'UTRs. PLoS Pathog. 2010, 6, e1000967. [Google Scholar] [CrossRef] [PubMed]
- Brest, P.; Lapaquette, P.; Souidi, M.; Lebrigand, K.; Cesaro, A.; Vouret-Craviari, V.; Mari, B.; Barbry, P.; Mosnier, J.F.; Hebuterne, X.; et al. A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn's disease. Nat. Genet. 2011, 43, 242–245. [Google Scholar] [CrossRef] [PubMed]
- Loeb, G.B.; Khan, A.A.; Canner, D.; Hiatt, J.B.; Shendure, J.; Darnell, R.B.; Leslie, C.S.; Rudensky, A.Y. Transcriptome-wide miR-155 binding map reveals widespread noncanonical microRNA targeting. Mol. Cell 2012, 48, 760–770. [Google Scholar] [CrossRef] [PubMed]
- Voight, B.F.; Kudaravalli, S.; Wen, X.; Pritchard, J.K. A map of recent positive selection in the human genome. PLoS Biol. 2006, 4, e72. [Google Scholar] [CrossRef] [PubMed]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, S.B.; Sammeth, M.; Gutierrez-Arcelus, M.; Lach, R.P.; Ingle, C.; Nisbett, J.; Guigo, R.; Dermitzakis, E.T. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature 2010, 464, 773–777. [Google Scholar] [CrossRef] [PubMed]
- Ng, P.C.; Henikoff, S. Predicting deleterious amino acid substitutions. Genome Res. 2001, 11, 863–874. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Qiu, C.; Cui, Q. A Large-Scale Analysis of the Relationship of Synonymous SNPs Changing MicroRNA Regulation with Functionality and Disease. Int. J. Mol. Sci. 2015, 16, 23545-23555. https://doi.org/10.3390/ijms161023545
Wang Y, Qiu C, Cui Q. A Large-Scale Analysis of the Relationship of Synonymous SNPs Changing MicroRNA Regulation with Functionality and Disease. International Journal of Molecular Sciences. 2015; 16(10):23545-23555. https://doi.org/10.3390/ijms161023545
Chicago/Turabian StyleWang, Yuchen, Chengxiang Qiu, and Qinghua Cui. 2015. "A Large-Scale Analysis of the Relationship of Synonymous SNPs Changing MicroRNA Regulation with Functionality and Disease" International Journal of Molecular Sciences 16, no. 10: 23545-23555. https://doi.org/10.3390/ijms161023545






