Systematic Analysis of the Maize PHD-Finger Gene Family Reveals a Subfamily Involved in Abiotic Stress Response
Abstract
:1. Introduction
2. Results
2.1. Identification of PHD Proteins in Maize
| Gene Name | Gene Identifier | Protein Size (aa) | 5′ End | 3′ End | Chromosme |
|---|---|---|---|---|---|
| ZmPHD1 | GRMZM5G862565 | 1579 | 100,335,533 | 100,348,389 | I |
| ZmPHD2 | GRMZM5G866423 | 287 | 209,536,012 | 209,538,518 | I |
| ZmPHD3 | GRMZM5G813111 | 252 | 276,110,228 | 276,115,825 | I |
| ZmPHD4 | AC225147.4 | 250 | 292,875,307 | 292,879,433 | I |
| ZmPHD5 | GRMZM2G403562 | 1900 | 144,629,199 | 144,637,545 | II |
| ZmPHD6 | GRMZM2G013936 | 751 | 155,881,115 | 15,590,458 | II |
| ZmPHD7 | GRMZM2G047316 | 241 | 158,280,766 | 158,284,014 | II |
| ZmPHD8 | GRMZM2G409224 | 1566 | 198,098,543 | 198,115,319 | II |
| ZmPHD9 | GRMZM2G181158 | 345 | 201,127,832 | 201,131,690 | II |
| ZmPHD10 | GRMZM2G158194 | 556 | 9,042,243 | 9,044,511 | III |
| ZmPHD11 | GRMZM2G085266 | 971 | 33,200,782 | 32,090,831 | III |
| ZmPHD12 | GRMZM2G069886 | 1214 | 114,434,585 | 114,444,762 | III |
| ZmPHD13 | GRMZM2G081350 | 818 | 133,742,782 | 133,749,539 | III |
| ZmPHD14 | GRMZM2G153087 | 257 | 135,907,092 | 135,912,402 | III |
| ZmPHD15 | GRMZM2G080917 | 257 | 170,106,279 | 170,110,892 | III |
| ZmPHD16 | GRMZM2G025703 | 173 | 171,031,966 | 171,035,433 | III |
| ZmPHD17 | GRMZM2G314546 | 1577 | 197,392,618 | 197,405,889 | III |
| ZmPHD18 | GRMZM2G068331 | 697 | 10,228,455 | 10,233,101 | IV |
| ZmPHD19 | GRMZM2G008259 | 172 | 11,389,157 | 11,413,875 | IV |
| ZmPHD20 | GRMZM2G059266 | 558 | 40,354,679 | 40,358,758 | IV |
| ZmPHD21 | GRMZM5G871463 | 72 | 79,000,726 | 79,005,980 | IV |
| ZmPHD22 | GRMZM2G107807 | 255 | 120,625,503 | 120,629,020 | IV |
| ZmPHD23 | GRMZM2G067019 | 418 | 136,070,311 | 136,101,261 | IV |
| ZmPHD24 | GRMZM2G385338 | 1812 | 234,187,863 | 234,200,009 | IV |
| ZmPHD25 | GRMZM2G391413 | 1322 | 239,318,755 | 239,331,611 | IV |
| ZmPHD26 | GRMZM2G110952 | 1166 | 2,576,103 | 2,593,650 | V |
| ZmPHD27 | GRMZM2G087482 | 1376 | 7,443,884 | 7,459,423 | V |
| ZmPHD28 | GRMZM2G466292 | 527 | 65,425,225 | 65,434,544 | V |
| ZmPHD29 | GRMZM2G466270 | 1290 | 65,442,955 | 65,456,381 | V |
| ZmPHD30 | GRMZM2G063864 | 255 | 174,409,725 | 174,413,665 | V |
| ZmPHD31 | GRMZM2G045544 | 727 | 203,646,189 | 203,650,857 | V |
| ZmPHD32 | GRMZM2G368206 | 1465 | 210,502,142 | 210,518,679 | V |
| ZmPHD33 | GRMZM2G039895 | 555 | 1,537,396 | 1,541,397 | VI |
| ZmPHD34 | GRMZM2G134214 | 849 | 54,775,354 | 54,785,293 | VI |
| ZmPHD35 | GRMZM2G103230 | 1147 | 58,263,970 | 58,269,763 | VI |
| ZmPHD36 | GRMZM2G473258 | 321 | 94,516,049 | 94,518,519 | VI |
| ZmPHD37 | GRMZM2G168249 | 808 | 107,094,897 | 107,103,424 | VI |
| ZmPHD38 | GRMZM2G330024 | 1108 | 109,391,739 | 109,405,354 | VI |
| ZmPHD39 | GRMZM2G148810 | 253 | 124,705,685 | 124,710,238 | VI |
| ZmPHD40 | GRMZM2G016817 | 256 | 147,643,515 | 147,650,880 | VI |
| ZmPHD41 | GRMZM2G412492 | 999 | 8,826,124 | 8,831,882 | VII |
| ZmPHD42 | GRMZM2G097726 | 219 | 13,348,616 | 13,356,443 | VII |
| ZmPHD43 | GRMZM2G013794 | 976 | 72,242,570 | 72,287,055 | VII |
| ZmPHD44 | GRMZM2G091265 | 212 | 105,907,436 | 105,911,988 | VII |
| ZmPHD45 | GRMZM2G316191 | 2379 | 150,159,272 | 150,178,742 | VII |
| ZmPHD46 | GRMZM2G128176 | 1712 | 171,564,760 | 171,575,198 | VII |
| ZmPHD47 | GRMZM2G434715 | 771 | 174,765,898 | 174,774,731 | VII |
| ZmPHD48 | GRMZM5G893976 | 241 | 15,274,043 | 15,278,560 | VIII |
| ZmPHD49 | GRMZM2G372928 | 759 | 25,354,740 | 25,391,185 | VIII |
| ZmPHD50 | GRMZM2G158918 | 256 | 103,454,290 | 103,460,620 | VIII |
| ZmPHD51 | GRMZM2G017142 | 253 | 125,536,101 | 125,539,093 | VIII |
| ZmPHD52 | GRMZM2G335720 | 836 | 137,142,057 | 137,164,518 | VIII |
| ZmPHD53 | GRMZM2G170412 | 870 | 147,718,464 | 147,724,348 | VIII |
| ZmPHD54 | GRMZM2G149587 | 381 | 156,906,971 | 156,912,759 | VIII |
| ZmPHD55 | GRMZM2G172001 | 255 | 164,744,187 | 164,748,517 | VIII |
| ZmPHD56 | GRMZM2G455243 | 852 | 12,345,652 | 12,367,654 | IX |
| ZmPHD57 | GRMZM2G156129 | 108 | 29,157,055 | 29,158,213 | IX |
| ZmPHD58 | GRMZM2G472428 | 1990 | 30,050,060 | 30,074,947 | IX |
| ZmPHD59 | GRMZM5G889372 | 868 | 136,475,636 | 136,480,652 | IX |
| ZmPHD60 | GRMZM2G178072 | 249 | 152,186,947 | 152,194,716 | IX |
| ZmPHD61 | GRMZM2G314661 | 1869 | 4,871,817 | 4,880,152 | X |
| ZmPHD62 | GRMZM2G365888 | 819 | 27,266,656 | 27,273,862 | X |
| ZmPHD63 | GRMZM2G115424 | 245 | 31,307,377 | 31,312,172 | X |
| ZmPHD64 | GRMZM2G404426 | 421 | 65,737,118 | 65,752,360 | X |
| ZmPHD65 | GRMZM2G156088 | 249 | 119,469,068 | 119,473,971 | X |
| ZmPHD66 | GRMZM2G050495 | 248 | 119,702,189 | 119,707,621 | X |
| ZmPHD67 | GRMZM2G038050 | 1339 | 149,547,787 | 149,558,325 | X |
2.2. Phylogenetic and Structural Analysis
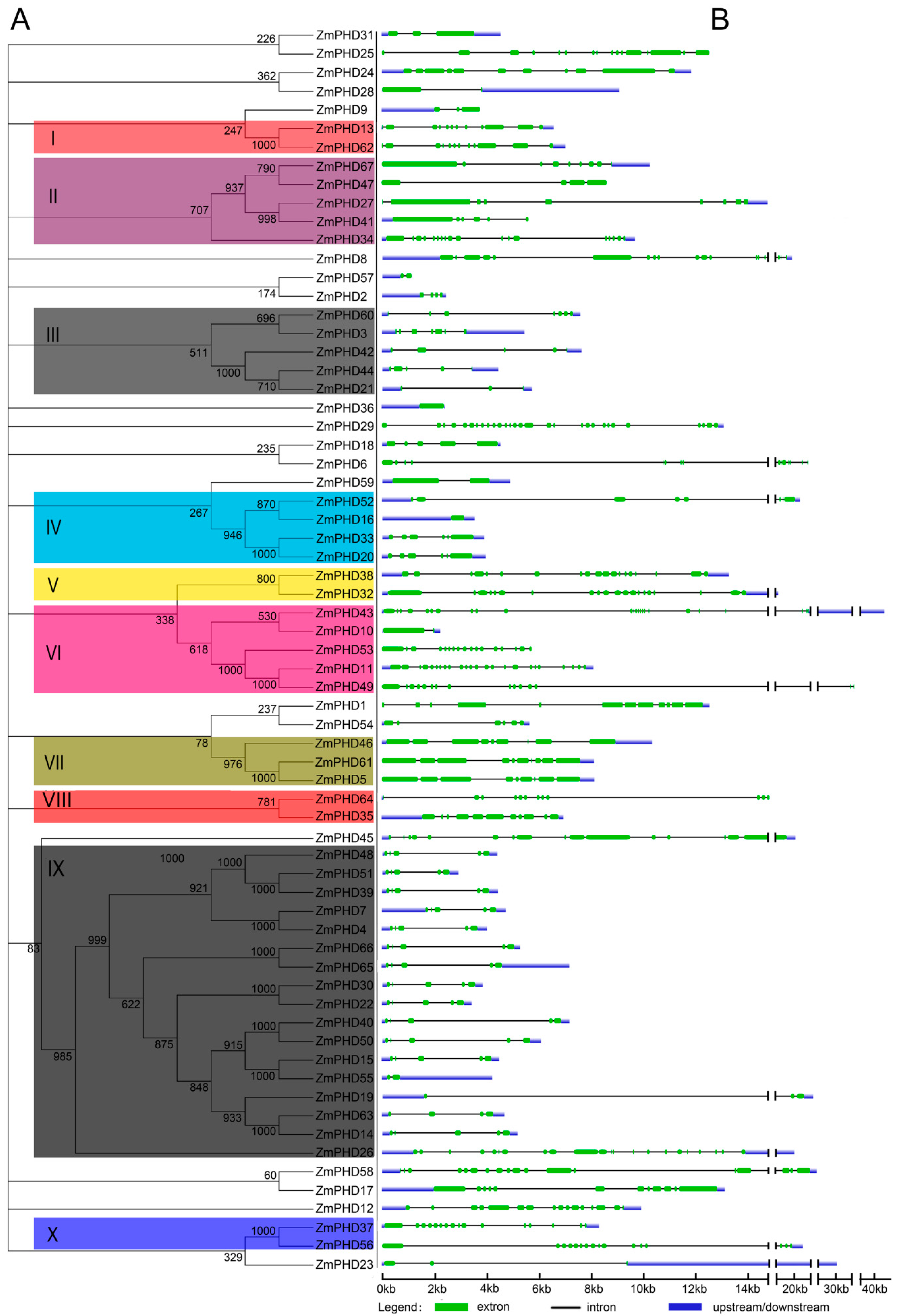
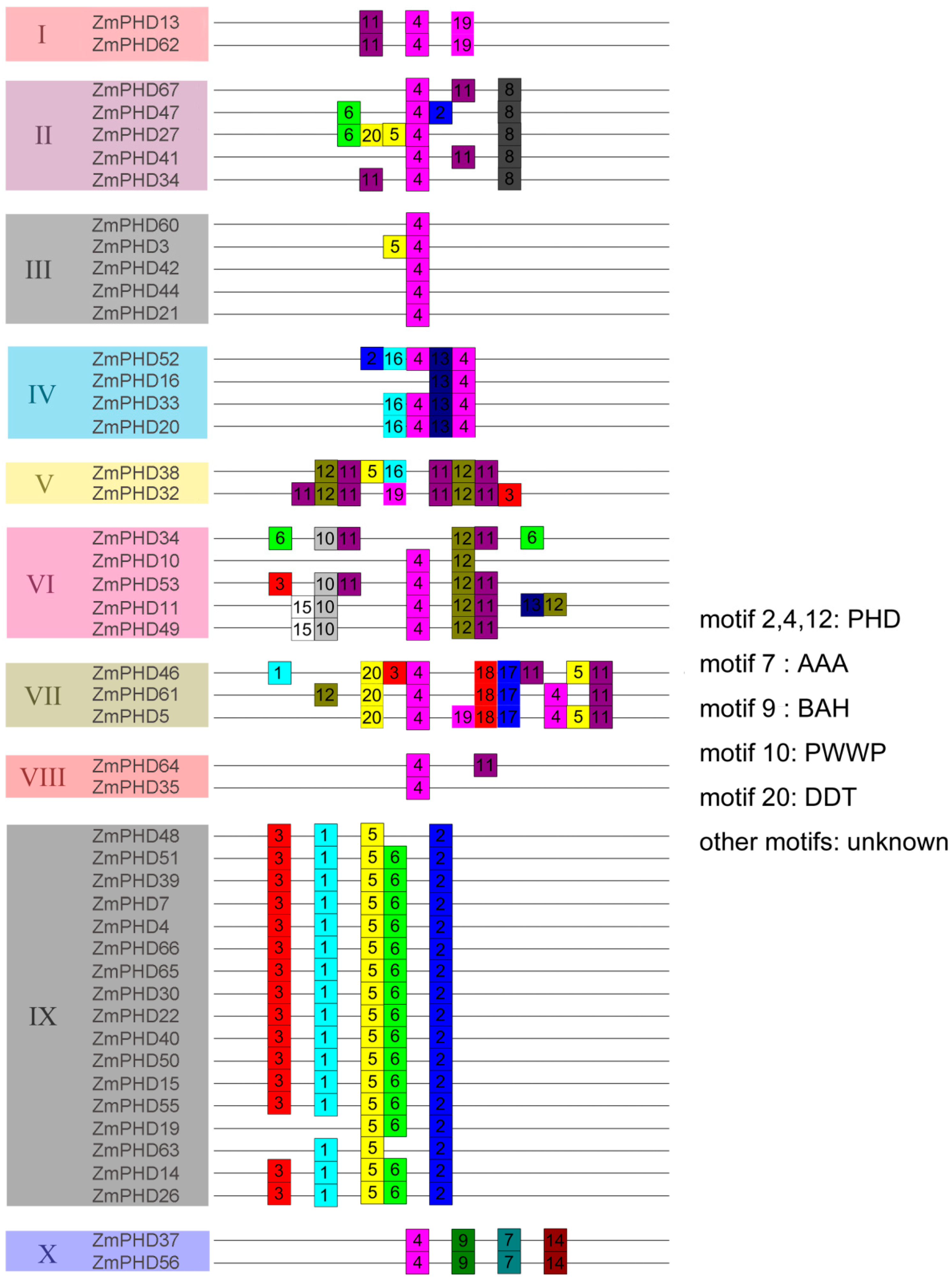
2.3. The Sequence Analysis of Maize PHD-Finger Domain and Motifs

2.4. Chromosomal Locations and Duplications of ZmPHDs
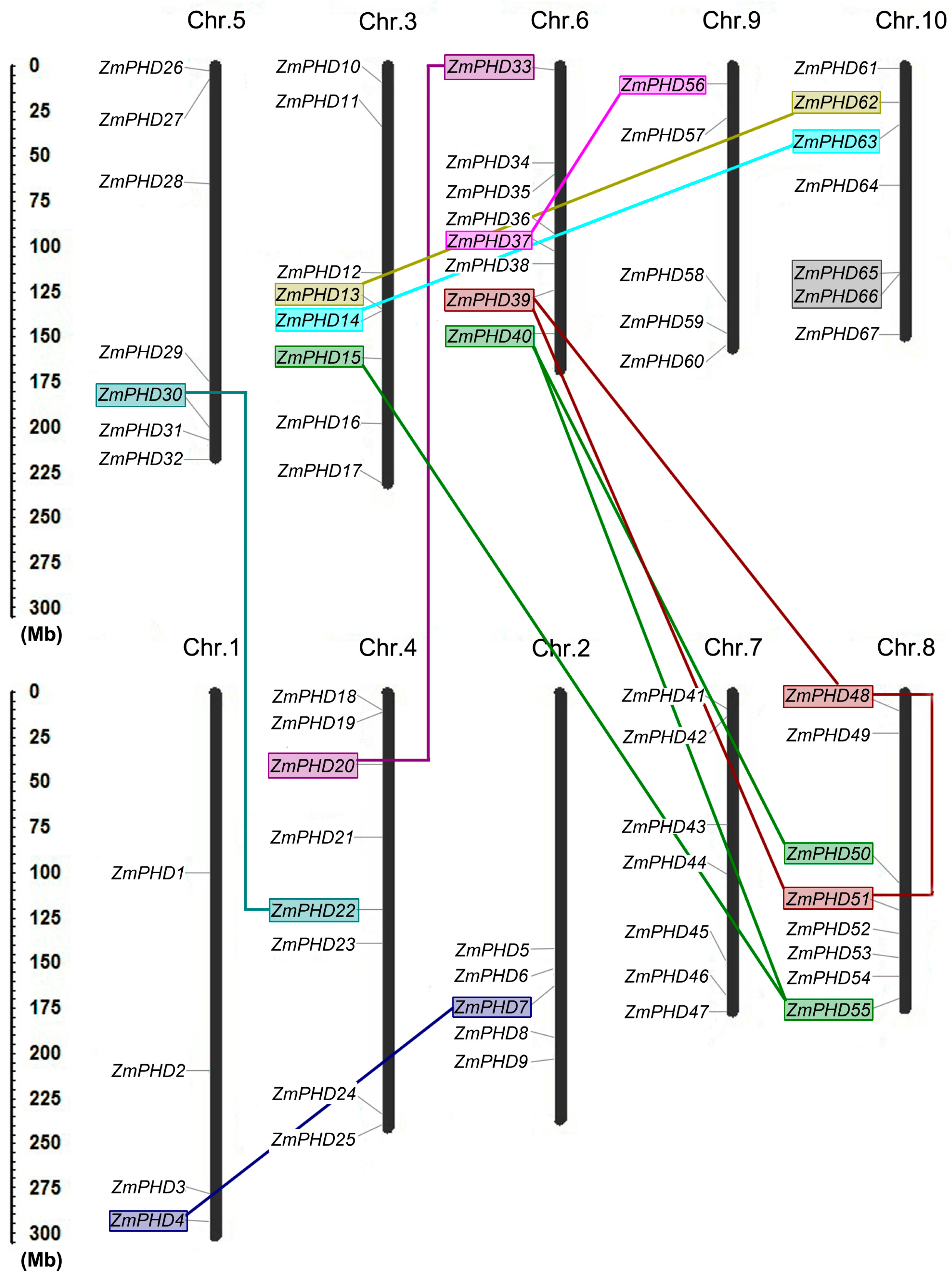
| Paralogous Pairs | Ks | Ka | Ka/Ks | Duplication Date (MY) | Duplicate Type |
|---|---|---|---|---|---|
| ZmPHD4/7 | 0.969 | 0.175 | 0.181 | 74.53 | segmental |
| ZmPHD13/62 | 0.148 | 0.031 | 0.209 | 16.07 | segmental |
| ZmPHD14/63 | 0.176 | 0.026 | 0.147 | 13.53 | segmental |
| ZmPHD15/55 | 0.686 | 0.436 | 0.635 | 52.76 | segmental |
| ZmPHD20/33 | 0.23 | 0.078 | 0.339 | 17.69 | segmental |
| ZmPHD22/30 | 0.151 | 0.019 | 0.125 | 11.61 | segmental |
| ZmPHD37/56 | 0.188 | 0.025 | 0.132 | 14.46 | segmental |
| ZmPHD40/50 | 0.131 | 0.02 | 0.152 | 10.07 | segmental |
| ZmPHD40/55 | 0.7 | 0.535 | 0.764 | 53.84 | segmental |
| ZmPHD48/39 | 0.657 | 0.112 | 0.17 | 50.53 | segmental |
| ZmPHD48/51 | 0.627 | 0.109 | 0.173 | 48.23 | segmental |
| ZmPHD51/39 | 0.236 | 0.02 | 0.084 | 18.15 | segmental |
| ZmPHD65/66 | 0.096 | 0.059 | 0.614 | 7.38 | tandem |
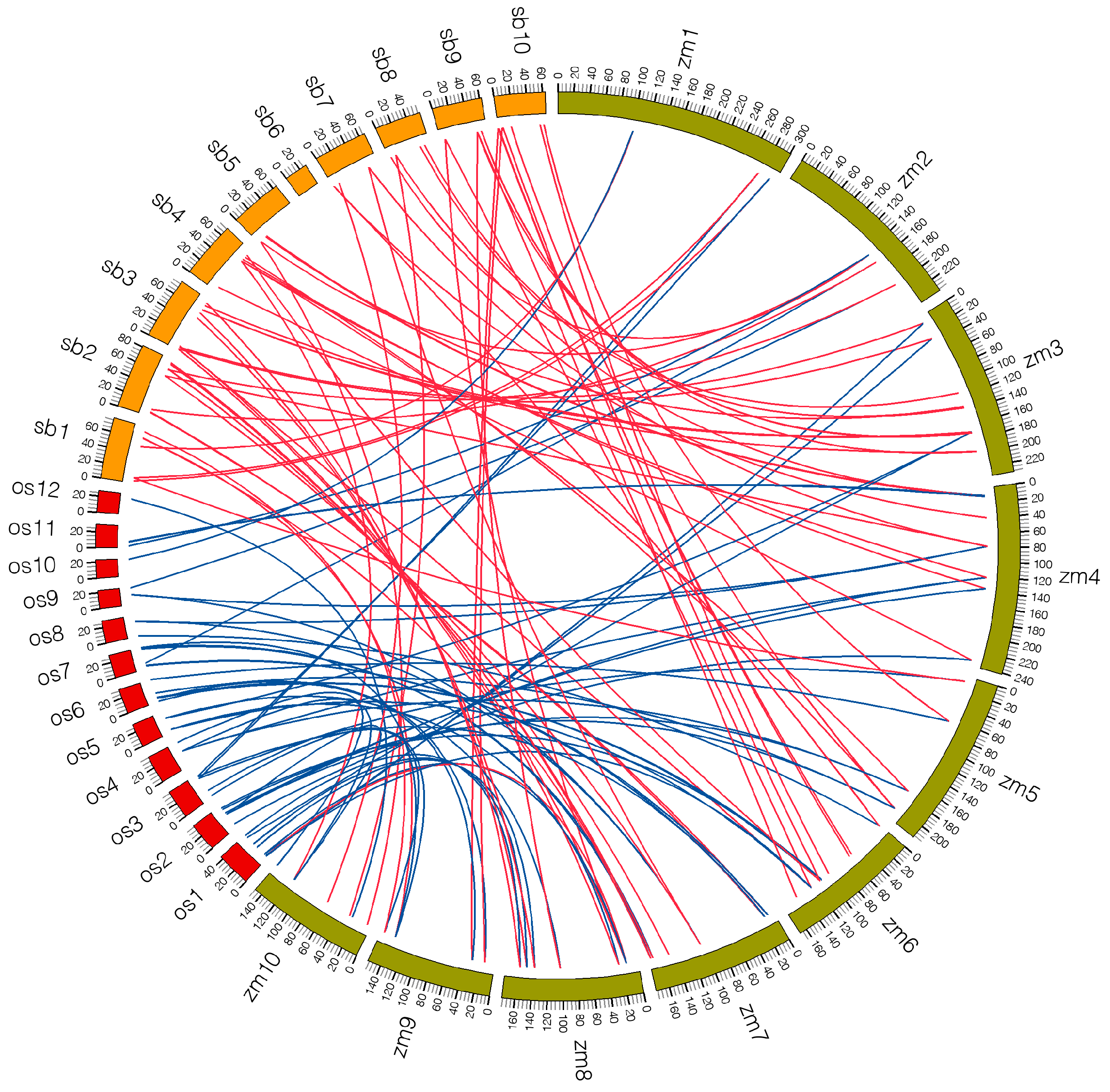
2.5. Microsynteny Analysis among Maize, Sorghum and Rice
2.6. Analysis of the Promoters of Potential Abiotic Stress-Responsive PHD Genes
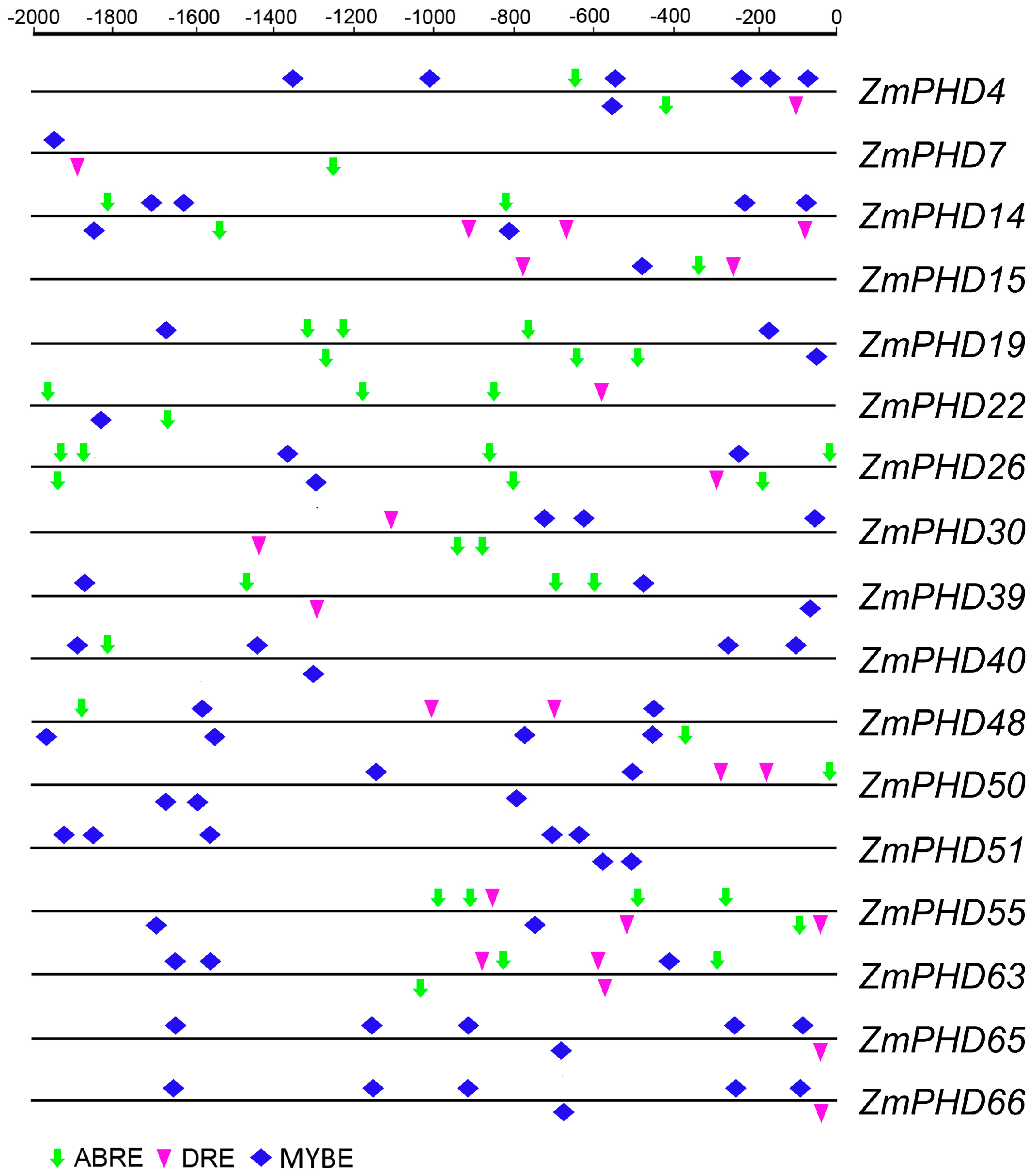
2.7. Microarray Expression Profiles of Maize PHD Genes

2.8. Expression Levels of Group IX PHD Genes in Response to Salt, PEG, and ABA Stress Condition
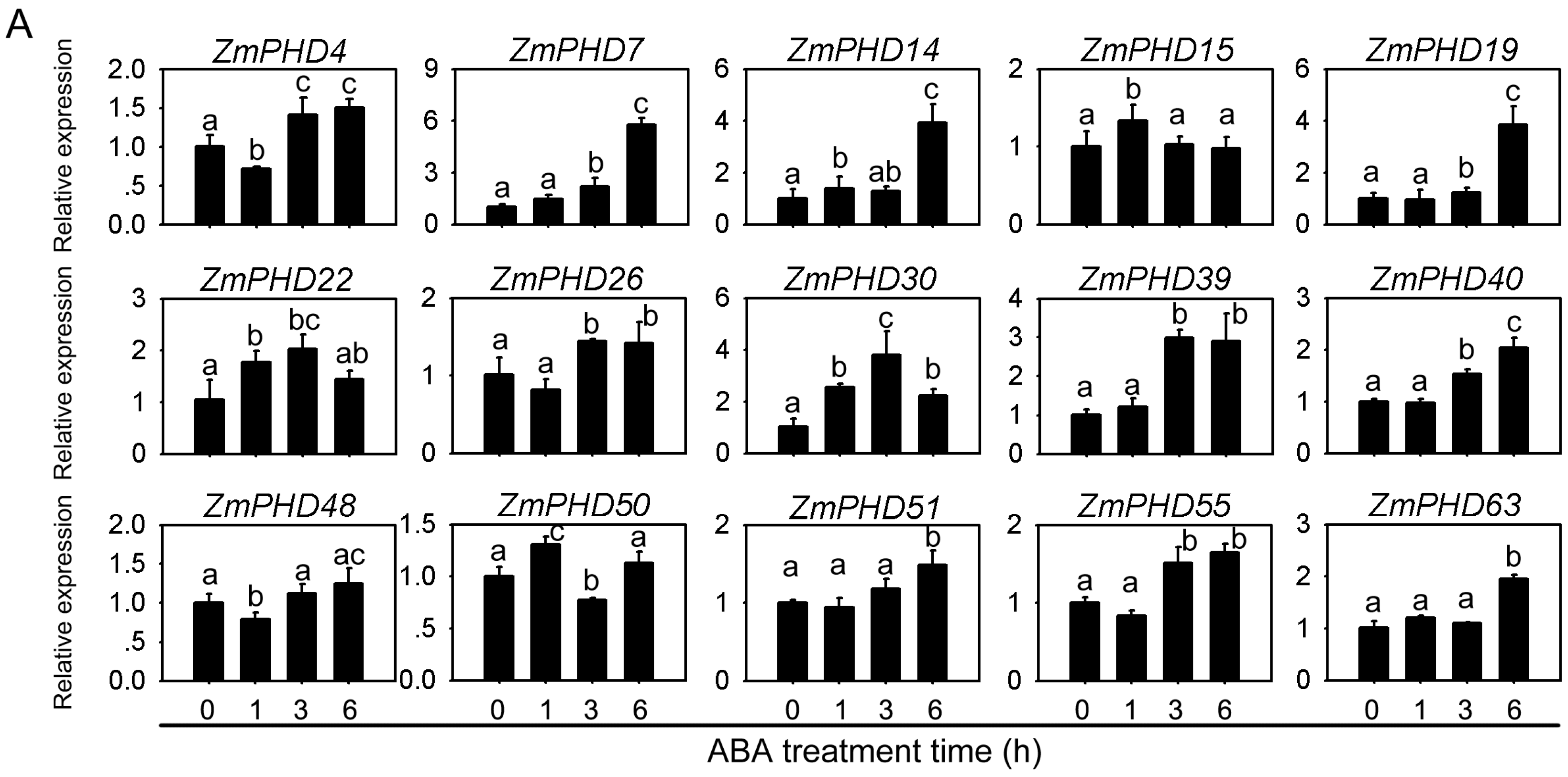
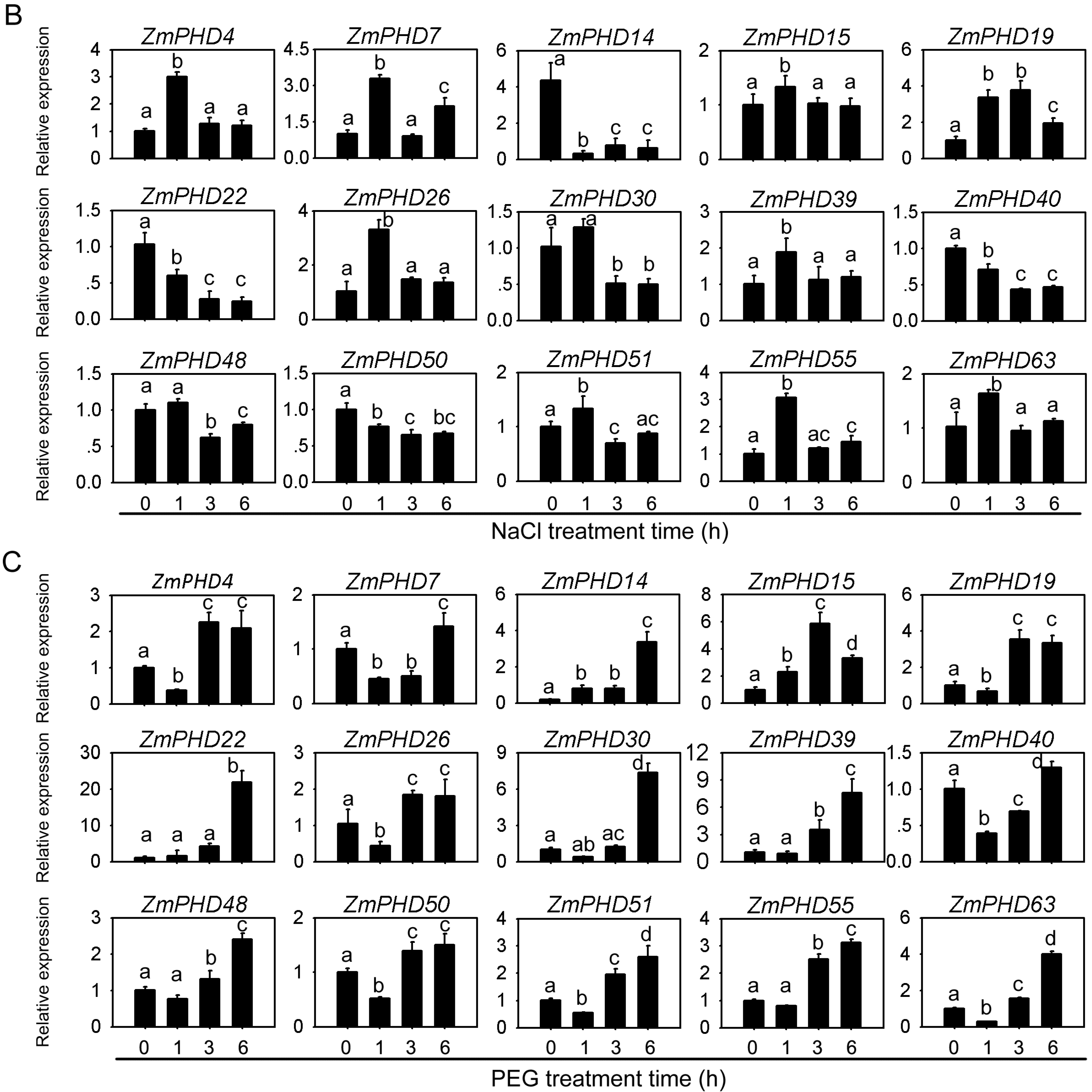
2.9. Subcellular Localization of ZmPHD14 and ZmPHD19
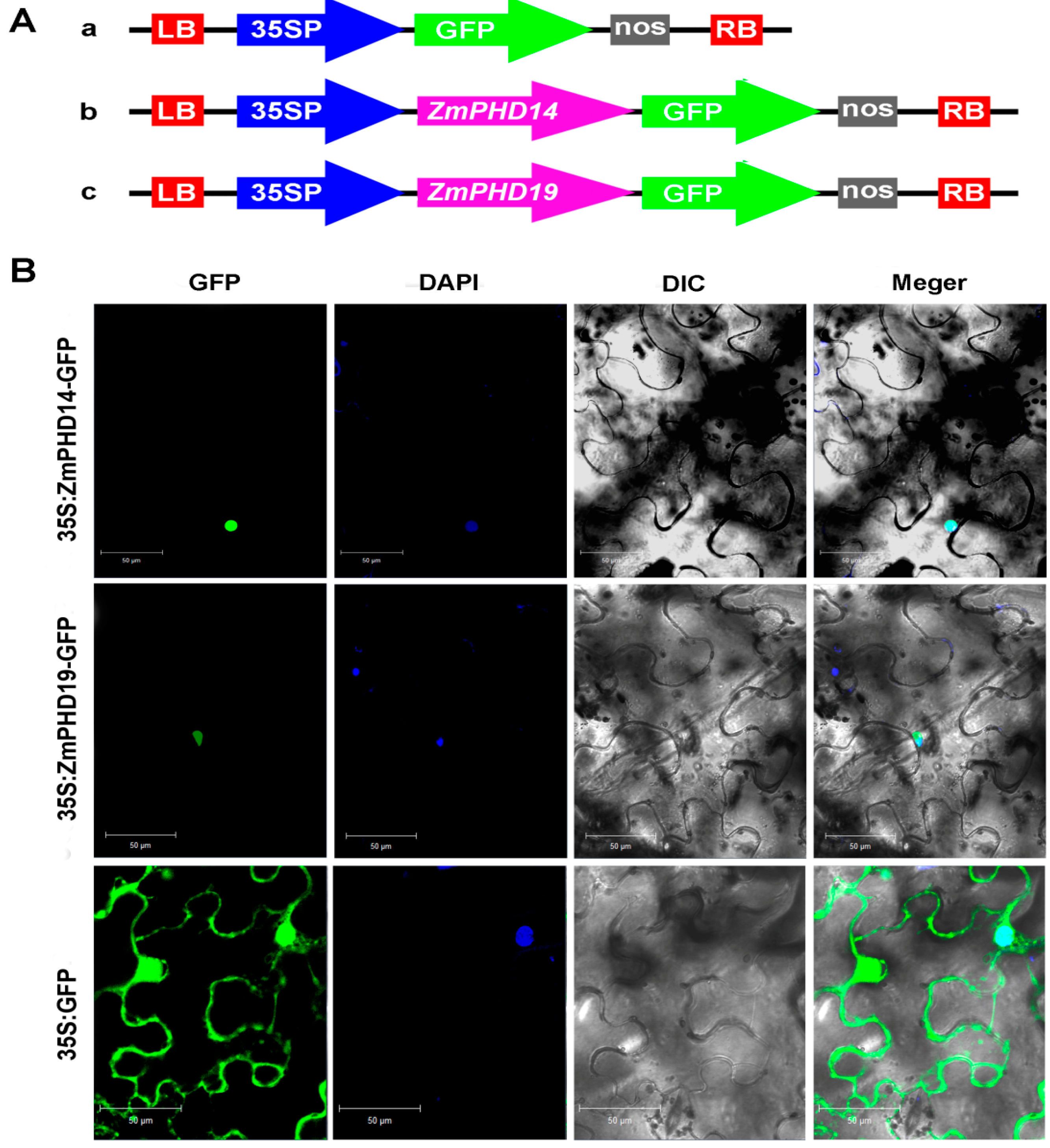
3. Discussion
4. Experimental Section
4.1. Collection and Classification of PHD-Finger Transcription Factors
4.2. Phylogenetic and Gene Structure Analysis
4.3. The Analysis of Chromosome Location and Gene Duplication
4.4. Interspecies Microsynteny Analysis
4.5. Cis-Elements in the Promoter Regions of Stress-Responsive PHD Class IX Genes
4.6. Microarray-Based Expression Analysis of Maize PHD Genes
4.7. Plant Materials and Stress Treatment
4.8. RNA Isolation and qRT-PCR Analysis
4.9. Determination of Subcellular Localization of ZmPHD14 and ZmPHD19
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Takatsuji, H. Zinc-finger transcription factors in plants. Cell. Mol. Life Sci. 1998, 54, 582–596. [Google Scholar] [CrossRef] [PubMed]
- Aasland, R.; Gibson, T.J.; Stewart, A.F. The PHD finger: Implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci. 1995, 20, 56–59. [Google Scholar] [CrossRef]
- Borden, K.L.; Freemont, P.S. The ring finger domain: A recent example of a sequence-structure family. Curr. Opin. Struct. Biol. 1996, 6, 395–401. [Google Scholar] [CrossRef]
- Townsley, F.M.; Thompson, B.; Bienz, M. Pygopus residues required for its binding to legless are critical for transcription and development. J. Biol. Chem. 2004, 279, 5177–5183. [Google Scholar] [CrossRef] [PubMed]
- Schindler, U.; Beckmann, H.; Cashmore, A.R. HAT3.1, a novel Arabidopsis homeodomain protein containing a conserved cysteine-rich region. Plant J. 1993, 4, 137–150. [Google Scholar] [CrossRef] [PubMed]
- Champagne, K.S.; Kutateladze, T.G. Structural insight into histone recognition by the ING PHD fingers. Curr. Drug Targets 2009, 10, 432–441. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.G.; Baetz, K.; Shi, X.; Walter, K.L.; MacDonald, V.E.; Wlodarski, M.J.; Gozani, O.; Hieter, P.; Howe, L. The Yng1p plant homeodomain finger is a methyl-histone binding module that recognizes lysine 4-methylated histone H3. Mol. Cell. Biol. 2006, 26, 7871–7879. [Google Scholar] [CrossRef] [PubMed]
- Wysocka, J.; Swigut, T.; Xiao, H.; Milne, T.A.; Kwon, S.Y.; Landry, J.; Kauer, M.; Tackett, A.J.; Chait, B.T.; Badenhorst, P. A PHD finger of nurf couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 2006, 442, 86–90. [Google Scholar] [CrossRef] [PubMed]
- Reddy, T.V.; Kaur, J.; Agashe, B.; Sundaresan, V.; Siddiqi, I. The duet gene is necessary for chromosome organization and progression during male meiosis in arabidopsis and encodes a PHD finger protein. Development 2003, 130, 5975–5987. [Google Scholar] [CrossRef] [PubMed]
- Woo, H.R.; Pontes, O.; Pikaard, C.S.; Richards, E.J. VIM1, a methylcytosine-binding protein required for centromeric heterochromatinization. Genes Dev. 2007, 21, 267–277. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Makaroff, C.A.; Ma, H. The Arabidopsis MALE MEIOCYTE DEATH1 gene encodes a PHD-finger protein that is required for male meiosis. Plant Cell 2003, 15, 1281–1295. [Google Scholar] [CrossRef] [PubMed]
- Barlow, J.H.; Faryabi, R.B.; Callén, E.; Wong, N.; Malhowski, A.; Chen, H.T.; Gutierrez-Cruz, G.; Sun, H.W.; McKinnon, P.; Wright, G. Identification of early replicating fragile sites that contribute to genome instability. Cell 2013, 152, 620–632. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Zhang, X.; Song, S.; Tian, C.; Yin, Y.; Xing, G.; He, F.; Zhang, L. Identification of UHRF1/2 as new N-methylpurine DNA glycosylase-interacting proteins. Biochem. Biophys. Res. Commun. 2013, 433, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Gao, Q.; Li, P.; Zhao, Q.; Zhang, J.; Li, J.; Koseki, H.; Wong, J. UHRF1 targets DNMT1 for DNA methylation through cooperative binding of hemi-methylated DNA and methylated H3K9. Nat. Commun. 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Saleh, A.; Alvarez-Venegas, R.; Yilmaz, M.; Le, O.; Hou, G.; Sadder, M.; Al-Abdallat, A.; Xia, Y.; Lu, G.; Ladunga, I. The highly similar arabidopsis homologs of trithorax ATX1 and ATX2 encode proteins with divergent biochemical functions. Plant Cell 2008, 20, 568–579. [Google Scholar] [CrossRef] [PubMed]
- Merkel, D.J.; Wells, S.B.; Hilburn, B.C.; Elazzouzi, F.; Pérez-Alvarado, G.C.; Lee, B.M. The C-terminal region of cytoplasmic polyadenylation element binding protein is a ZZ domain with potential for protein–protein interactions. J. Mol. Biol. 2013, 425, 2015–2026. [Google Scholar] [CrossRef] [PubMed]
- Baker, L.A.; Allis, C.D.; Wang, G.G. PHD fingers in human diseases: Disorders arising from misinterpreting epigenetic marks. Mutat. Res./Fundam. Mol. Mech. Mutagen. 2008, 647, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Geng, Y.; Gao, Y.; Ju, H.; Yan, F. Diagnostic and prognostic value of plasma and tissue ubiquitin-like, containing PHD and ring finger domains 1 in breast cancer patients. Cancer Sci. 2013, 104, 194–199. [Google Scholar] [CrossRef] [PubMed]
- Coscoy, L.; Ganem, D. PHD domains and E3 ubiquitin ligases: Viruses make the connection. Trends Cell Biol. 2003, 13, 7–12. [Google Scholar] [CrossRef]
- Gozani, O.; Karuman, P.; Jones, D.R.; Ivanov, D.; Cha, J.; Lugovskoy, A.A.; Baird, C.L.; Zhu, H.; Field, S.J.; Lessnick, S.L. The PHD finger of the chromatin-associated protein ING2 functions as a nuclear phosphoinositide receptor. Cell 2003, 114, 99–111. [Google Scholar] [CrossRef]
- Wei, W.; Huang, J.; Hao, Y.J.; Zou, H.F.; Wang, H.W.; Zhao, J.Y.; Liu, X.Y.; Zhang, W.K.; Ma, B.; Zhang, J.S. Soybean GMPHD-type transcription regulators improve stress tolerance in transgenic Arabidopsis plants. PLoS ONE 2009, 4, e7209. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhou, Y.; Jiang, H.; Li, X.; Gan, D.; Peng, X.; Zhu, S.; Cheng, B. Systematic analysis of sequences and expression patterns of drought-responsive members of the HD-Zip gene family in maize. PLoS ONE 2011, 6, e28488. [Google Scholar]
- Schnable, P.S.; Ware, D.; Fulton, R.S.; Stein, J.C.; Wei, F.; Pasternak, S.; Liang, C.; Zhang, J.; Fulton, L.; Graves, T.A. The B73 maize genome: Complexity, diversity, and dynamics. Science 2009, 326, 1112–1115. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Zhao, Y.; Cao, J.; Zhang, W.; Jiang, H.; Li, X.; Ma, Q.; Zhu, S.; Cheng, B. CCCH-type zinc finger family in maize: Genome-wide identification, classification and expression profiling under abscisic acid and drought treatments. PLoS ONE 2012, 7, e40120. [Google Scholar] [CrossRef] [PubMed]
- Capili, A.D.; Schultz, D.C.; Rauscher, F.J.; Borden, K.L. Solution structure of the PHD domain from the KAP-1 corepressor: Structural determinants for PHD, RING and LIM zinc-binding domains. EMBO J. 2001, 20, 165–177. [Google Scholar] [CrossRef] [PubMed]
- Callebaut, I.; Courvalin, J.C.; Mornon, J.P. The BAH (bromo-adjacent homology) domain: A link between DNA methylation, replication and transcriptional regulation. FEBS Lett. 1999, 446, 189–193. [Google Scholar] [CrossRef]
- Wang, Y.; Reddy, B.; Thompson, J.; Wang, H.; Noma, K.I.; Yates, J.R.; Jia, S. Regulation of Set9-mediated H4K20 methylation by a PWWP domain protein. Mol. Cell 2009, 33, 428–437. [Google Scholar] [CrossRef] [PubMed]
- Qiu, C.; Sawada, K.; Zhang, X.; Cheng, X. The PWWP domain of mammalian DNA methyltransferase Dnmt3b defines a new family of DNA-binding folds. Nat. Struct. Mol. Biol. 2002, 9, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaut, B.S.; Morton, B.R.; McCaig, B.C.; Clegg, M.T. Substitution rate comparisons between grasses and palms: Synonymous rate differences at the nuclear gene Adh parallel rate differences at the plastid gene rbcL. Proc. Natl. Acad. Sci. USA 1996, 93, 10274–10279. [Google Scholar] [CrossRef] [PubMed]
- Swigoňová, Z.; Lai, J.; Ma, J.; Ramakrishna, W.; Llaca, V.; Bennetzen, J.L.; Messing, J. Close split of sorghum and maize genome progenitors. Genome Res. 2004, 14, 1916–1923. [Google Scholar] [CrossRef] [PubMed]
- Narusaka, Y.; Nakashima, K.; Shinwari, Z.K.; Sakuma, Y.; Furihata, T.; Abe, H.; Narusaka, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 2003, 34, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Goto, E.; Ishido, S.; Sato, Y.; Ohgimoto, S.; Ohgimoto, K.; Nagano-Fujii, M.; Hotta, H. c-MIR, a human E3 ubiquitin ligase, is a functional homolog of herpesvirus proteins MIR1 and MIR2 and has similar activity. J. Biol. Chem. 2003, 278, 14657–14668. [Google Scholar] [CrossRef] [PubMed]
- Mansouri, M.; Bartee, E.; Gouveia, K.; Hovey Nerenberg, B.T.; Barrett, J.; Thomas, L.; Thomas, G.; Mcfadden, G.; Früh, K. The PHD/LAP-domain protein M153R of myxomavirus is a ubiquitin ligase that induces the rapid internalization and lysosomal destruction of CD4. J. Virol. 2003, 77, 1427–1440. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Liu, Q.P.; Xue, Q.Z. Comparative phylogenetic analysis of the rice and Arabidopsis PHDfinger proteins. Acta Genet. Sin. 2004, 31, 1284–1293. [Google Scholar] [PubMed]
- Moore, R.C.; Purugganan, M.D. The early stages of duplicate gene evolution. Proc. Natl. Acad. Sci. USA 2003, 100, 15682–15687. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.X.; Jiang, H.Y.; Chu, Z.X.; Tang, X.L.; Zhu, S.W.; Cheng, B.J. Genome-wide identification, classification and analysis of heat shock transcription factor family in maize. BMC Genom. 2011, 12. [Google Scholar] [CrossRef] [PubMed]
- Mainali, H.R.; Chapman, P.; Dhaubhadel, S. Genome-wide analysis of Cyclophilin gene family in soybean (Glycine max). BMC Plant Biol. 2014, 14. [Google Scholar] [CrossRef] [PubMed]
- Dou, L.; Zhang, X.; Pang, C.; Song, M.; Wei, H.; Fan, S.; Yu, S. Genome-wide analysis of the WRKY gene family in cotton. Mol. Genet. Genom. 2014, 289, 1103–1121. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, D.; Wang, L.; Li, D. Genome-wide analysis of mitogen-activated protein kinase gene family in maize. Plant Mol. Biol. Rep. 2013, 31, 1446–1460. [Google Scholar] [CrossRef]
- Van de Peer, Y.; Maere, S.; Meyer, A. The evolutionary significance of ancient genome duplications. Nat. Rev. Genet. 2009, 10, 725–732. [Google Scholar] [CrossRef] [PubMed]
- Maere, S.; de Bodt, S.; Raes, J.; Casneuf, T.; van Montagu, M.; Kuiper, M.; van de Peer, Y. Modeling gene and genome duplications in eukaryotes. Proc. Natl. Acad. Sci. USA 2005, 102, 5454–5459. [Google Scholar] [CrossRef] [PubMed]
- Lai, J.; Ma, J.; Swigoňová, Z.; Ramakrishna, W.; Linton, E.; Llaca, V.; Tanyolac, B.; Park, Y.J.; Jeong, O.Y.; Bennetzen, J.L. Gene loss and movement in the maize genome. Genome Res. 2004, 14, 1924–1931. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Wang, Q.; Sun, R.; Xie, F.; Jones, D.C.; Zhang, B. Genome-wide identification and expression analysis of TCP transcription factors in gossypium raimondii. Sci. Rep. 2014, 4. [Google Scholar] [CrossRef] [PubMed]
- Lynch, M.; Conery, J.S. The evolutionary fate and consequences of duplicate genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, Q.; Zhao, Y.; Han, G.; Zhu, S. Systematic analysis of maize Class III peroxidase gene family reveals a conserved subfamily involved in abiotic stress response. Gene 2015, 566, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Blanc, G.; Wolfe, K.H. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 2004, 16, 1679–1691. [Google Scholar] [CrossRef] [PubMed]
- Zong, X.J.; Li, D.P.; Gu, L.K.; Li, D.Q.; Liu, L.X.; Hu, X.L. Abscisic acid and hydrogen peroxide induce a novel maize group C MAP kinase gene, ZmMPK7, which is responsible for the removal of reactive oxygen species. Planta 2008, 229, 485–495. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.; Dai, X.; Zhang, W.H. A R2R3-type myb gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J. Exp. Bot. 2012, 63, 2541–2556. [Google Scholar] [CrossRef]
- Bienz, M. The PHD finger, a nuclear protein-interaction domain. Trends Biochem. Sci. 2006, 31, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The clustal_x windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [PubMed]
- Clewley, J.P.; Arnold, C. Megalign. In Sequence Data Analysis Guidebook; Springer: New York, NY, USA, 1997; pp. 119–129. [Google Scholar]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Yang, S.; Zhang, X.; Yue, J.X.; Tian, D.; Chen, J.Q. Recent duplications dominate NBS-encoding gene expansion in two woody species. Mol. Genet. Genom. 2008, 280, 187–198. [Google Scholar]
- Zhao, P.; Li, Q.; Li, J.; Wang, L.; Ren, Z. Genome-wide identification and characterization of R2R3MYB family in solanum lycopersicum. Mol. Genet. Genom. 2014, 289, 1183–1207. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Sánchez-DelBarrio, J.C.; Messeguer, X.; Rozas, R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 2003, 19, 2496–2497. [Google Scholar] [CrossRef] [PubMed]
- Quraishi, U.M.; Abrouk, M.; Murat, F.; Pont, C.; Foucrier, S.; Desmaizieres, G.; Confolent, C.; Rivière, N.; Charmet, G.; Paux, E. Cross-genome map based dissection of a nitrogen use efficiency ortho-metaqtl in bread wheat unravels concerted cereal genome evolution. Plant J. 2011, 65, 745–756. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H. MCScanx: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- Higo, K.; Ugawa, Y.; Iwamoto, M.; Korenaga, T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999, 27, 297–300. [Google Scholar] [CrossRef] [PubMed]
- Sekhon, R.S.; Briskine, R.; Hirsch, C.N.; Myers, C.L.; Springer, N.M.; Buell, C.R.; de Leon, N.; Kaeppler, S.M. Maize gene atlas developed by rna sequencing and comparative evaluation of transcriptomes based on RNA sequencing and microarrays. PLoS ONE 2013, 8, e61005. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Gao, C.; Zheng, X.; Han, B. Identification of OsbZIP72 as a positive regulator of ABA response and drought tolerance in rice. Planta 2009, 229, 605–615. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Huang, Y.; Xiong, L. Characterization of stress-responsive CIPK genes in rice for stress tolerance improvement. Plant Physiol. 2007, 144, 1416–1428. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative ct method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Chung, E.; Seong, E.; Kim, Y.C.; Chung, E.J.; Oh, S.K.; Lee, S.; Park, J.M.; Joung, Y.H.; Choi, D. А method of high frequency virus induced gene silencing in Chili pepper (Capsicum annuum L. Bukang). Mol. Cells 2004, 17, 377–380. [Google Scholar] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Q.; Liu, J.; Wang, Y.; Zhao, Y.; Jiang, H.; Cheng, B. Systematic Analysis of the Maize PHD-Finger Gene Family Reveals a Subfamily Involved in Abiotic Stress Response. Int. J. Mol. Sci. 2015, 16, 23517-23544. https://doi.org/10.3390/ijms161023517
Wang Q, Liu J, Wang Y, Zhao Y, Jiang H, Cheng B. Systematic Analysis of the Maize PHD-Finger Gene Family Reveals a Subfamily Involved in Abiotic Stress Response. International Journal of Molecular Sciences. 2015; 16(10):23517-23544. https://doi.org/10.3390/ijms161023517
Chicago/Turabian StyleWang, Qianqian, Jinyang Liu, Yu Wang, Yang Zhao, Haiyang Jiang, and Beijiu Cheng. 2015. "Systematic Analysis of the Maize PHD-Finger Gene Family Reveals a Subfamily Involved in Abiotic Stress Response" International Journal of Molecular Sciences 16, no. 10: 23517-23544. https://doi.org/10.3390/ijms161023517





