Prediction of Mature MicroRNA and Piwi-Interacting RNA without a Genome Reference or Precursors
Abstract
:1. Introduction
2. Results and Discussion
2.1. Feature Selection
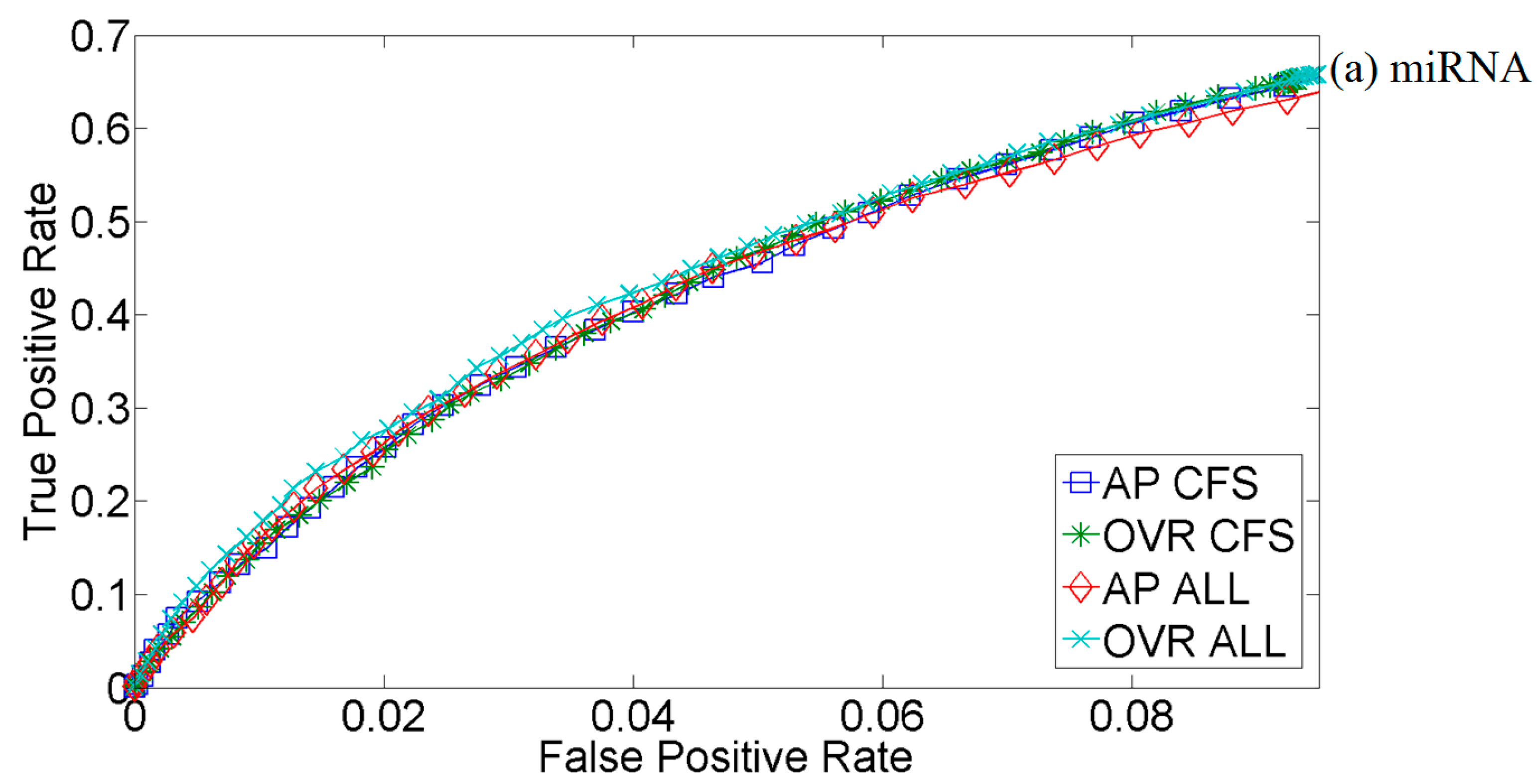
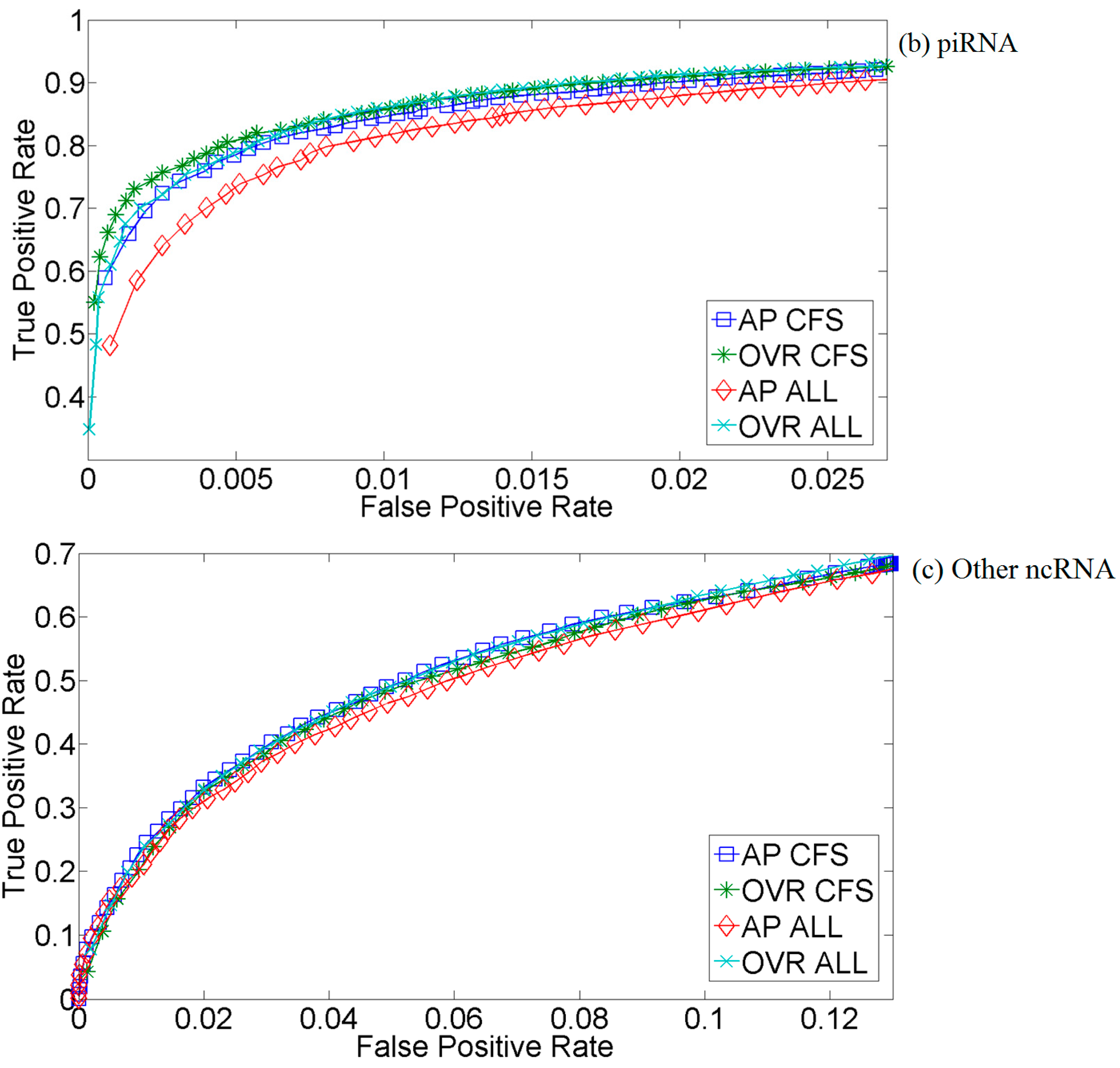
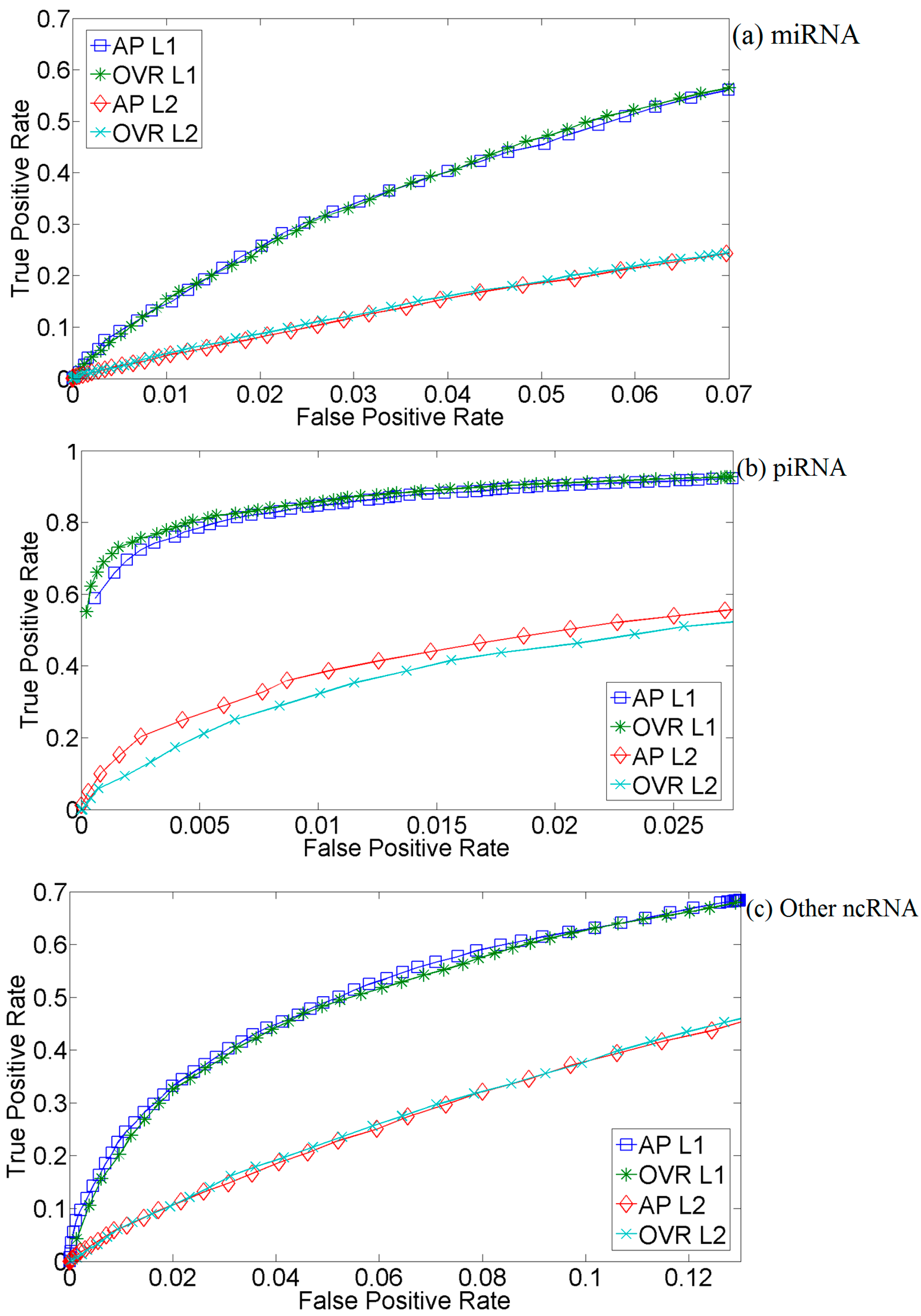
2.2. Kernel Selection
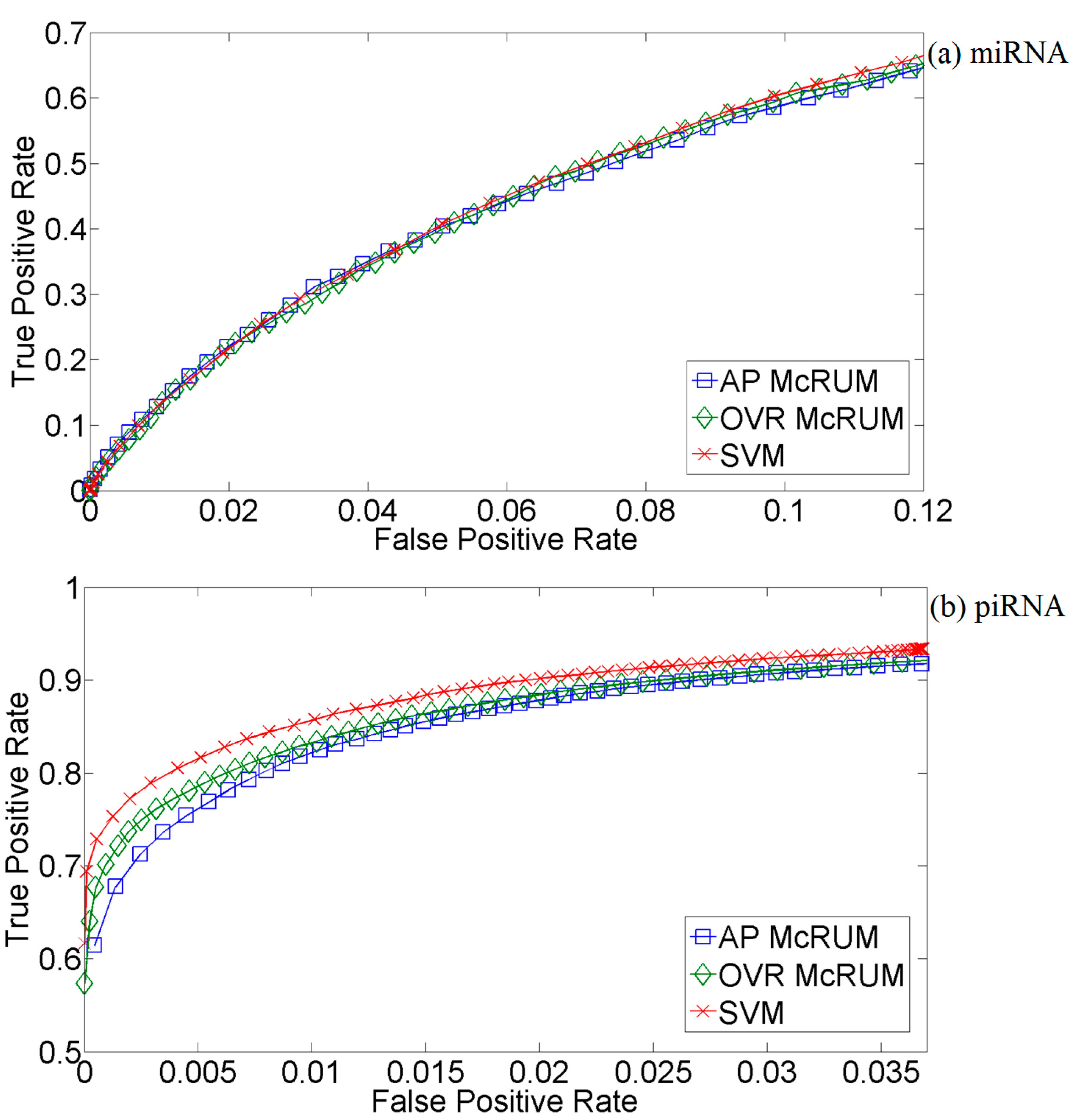
2.3. Comparison with SVM and piRNApredictor Using the Hold-Out Test Set
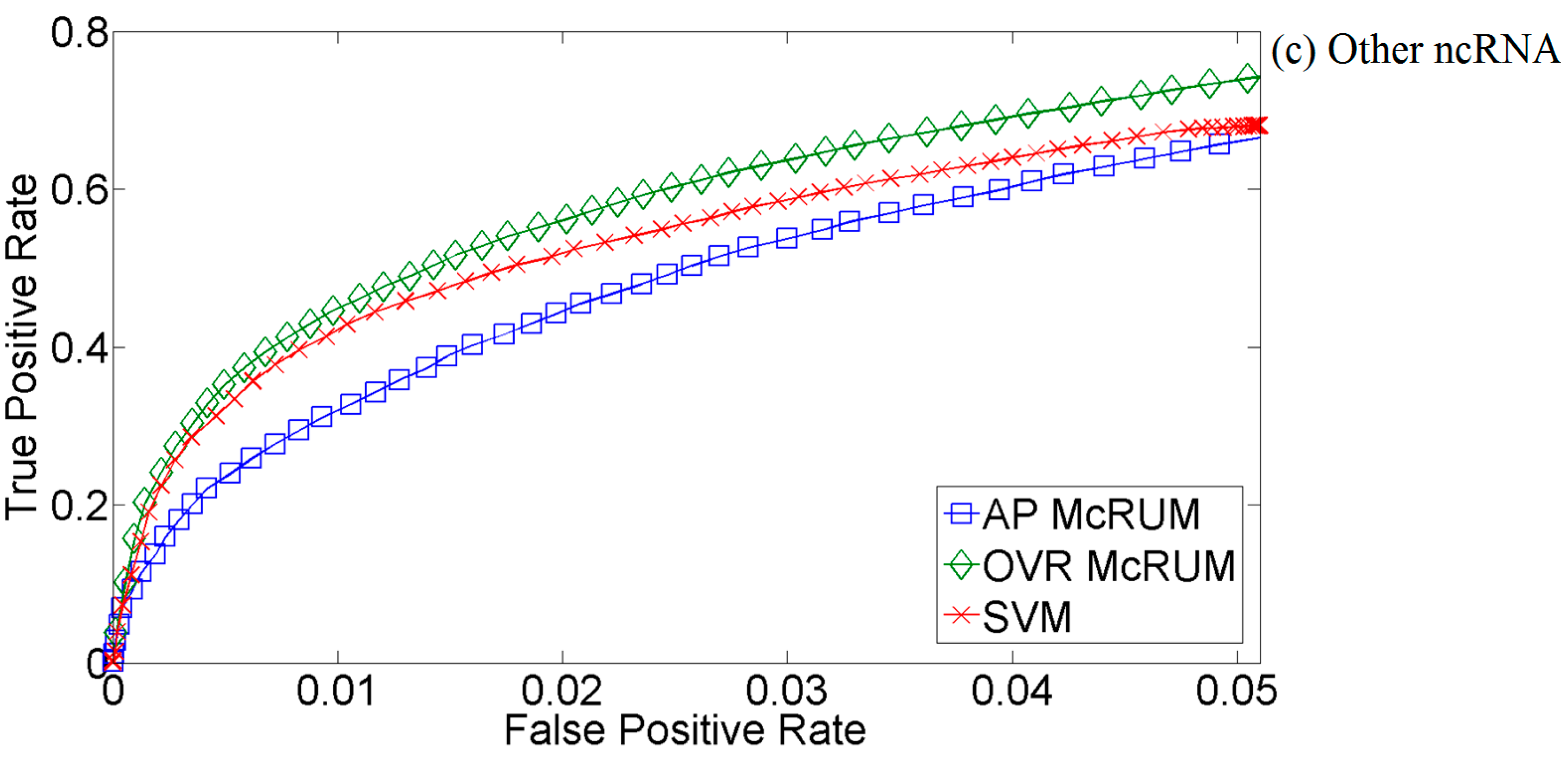
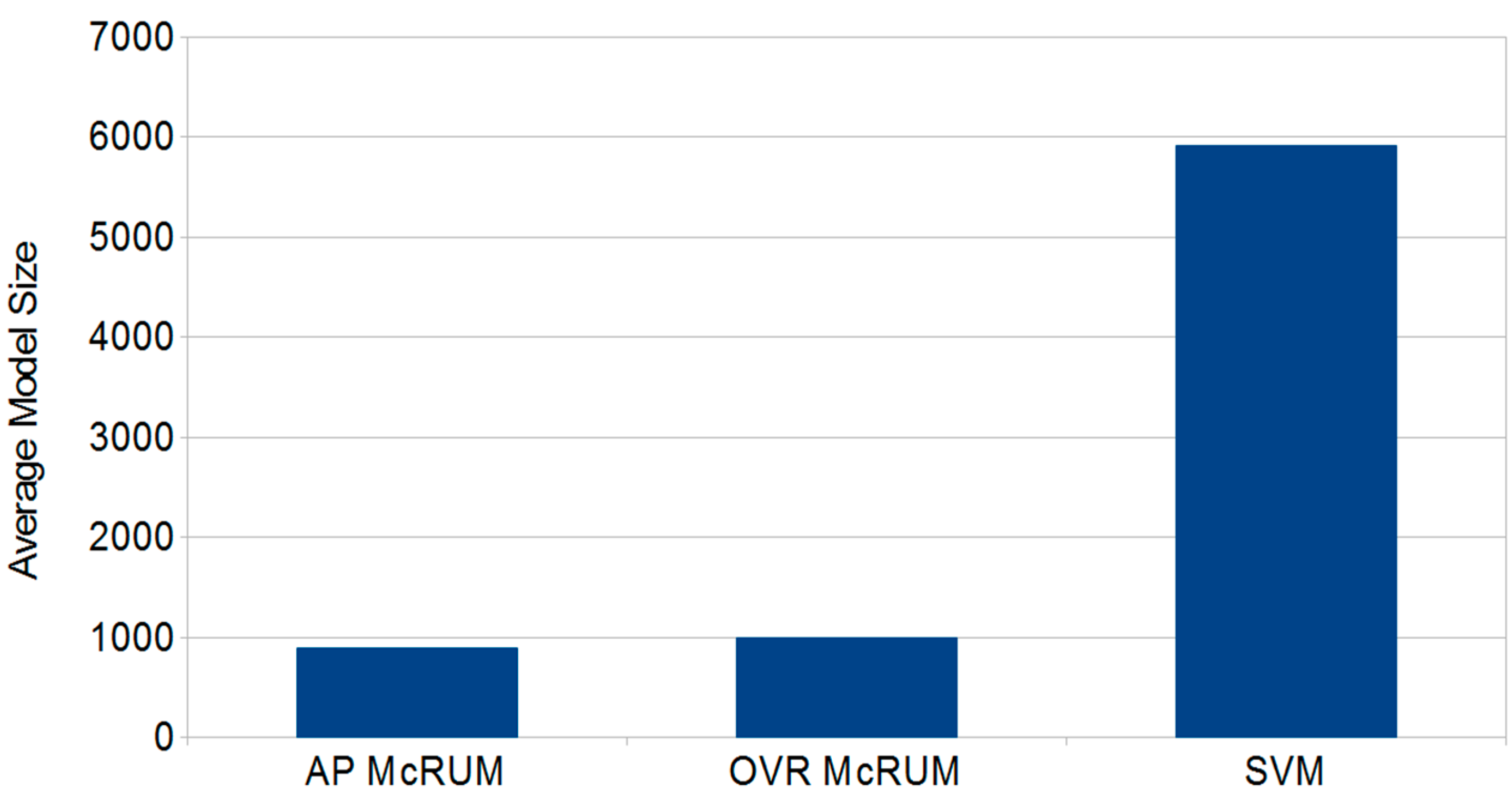
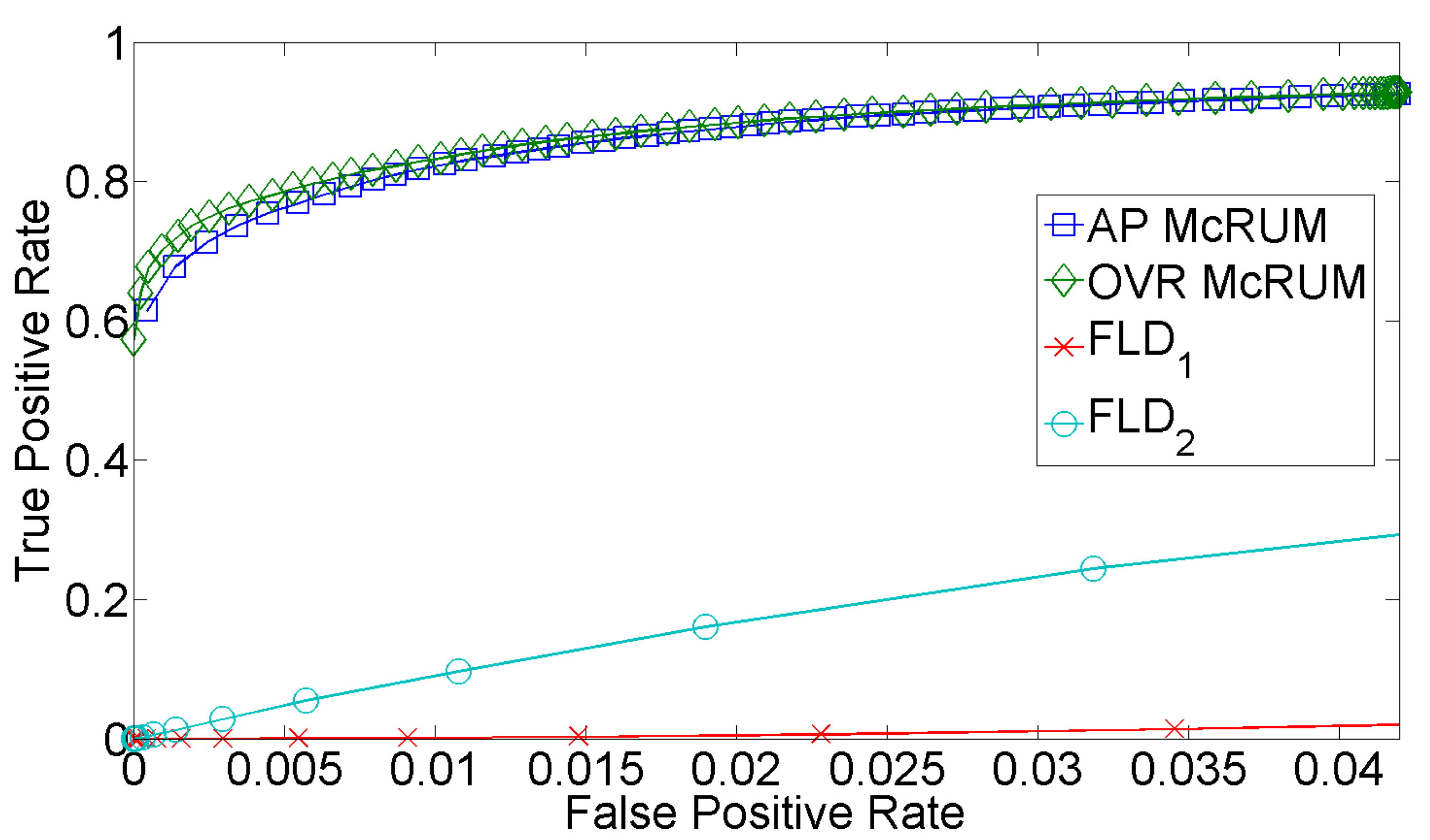
2.4. Comparison with miRPlex Using Experimental Datasets

3. Experimental Section
3.1. Classification Methods
3.2. Gaussian Kernels
3.3. Datasets
| Dataset | Species | Library Size | # of Unique Sequences | # of Filtered Sequences |
|---|---|---|---|---|
| GSM297747 | Caenorhabditis elegans | 5 million | 371,937 | 1353 |
| GSM317268 | Locusta migratoria | 1 million | 228,526 | 2091 |
| GSM317269 | Locusta migratoria | 2 million | 429,041 | 1668 |
| GSM609220 | Drosophila melanogaster | 14 million | 78,236 | 1349 |
3.4. Features
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Min, H.; Yoon, S. Got target?: Computational methods for microRNA target prediction and their extension. Exp. Mol. Med. 2010, 42, 233–244. [Google Scholar]
- Di Leva, G.; Croce, C.M. miRNA profiling of cancer. Curr. Opin. Genet. Dev. 2013, 23, 3–11. [Google Scholar]
- Castellano, L.; Stebbing, J. Deep sequencing of small RNAs identifies canonical and non-canonical miRNA and endogenous siRNAs in mammalian somatic tissues. Nucleic Acids Res. 2013, 41, 3339–3351. [Google Scholar] [CrossRef] [PubMed]
- Seto, A.G.; Kingston, R.E.; Lau, N.C. The coming of age for piwi proteins. Mol. Cell 2007, 26, 603–609. [Google Scholar] [CrossRef] [PubMed]
- Mani, S.R.; Juliano, C.E. Untangling the web: The diverse functions of the PIWI/piRNA pathway. Mol. Reprod. Dev. 2013, 80, 632–664. [Google Scholar] [CrossRef] [PubMed]
- Sato, K.; Siomi, M.C. Piwi-interacting RNAs: Biological functions and biogenesis. Essays Biochem. 2013, 54, 39–52. [Google Scholar] [CrossRef] [PubMed]
- Rajasethupathy, P.; Antonov, I.; Sheridan, R.; Frey, S.; Sander, C.; Tuschl, T.; Kandel, E.R. A role for neuronal piRNAs in the epigenetic control of memory-related synaptic plasticity. Cell 2012, 149, 693–707. [Google Scholar] [CrossRef] [PubMed]
- Luteijn, M.; Ketting, R. Piwi-interacting RNAs: From generation to transgenerational epigenetics. Nat. Rev. Genet. 2013, 14, 523–534. [Google Scholar] [CrossRef] [PubMed]
- Hess, A.M.; Prasad, A.N.; Ptitsyn, A.; Ebel, G.D.; Olson, K.E.; Barbacioru, C.; Monighetti, C.; Campbell, C.L. Small RNA profiling of Dengue virus-mosquito interactions implicates the PIWI RNA pathway in anti-viral defense. BMC Microbiol. 2011, 11. [Google Scholar] [CrossRef] [PubMed]
- Moxon, S.; Schwach, F.; Dalmay, T.; MacLean, D.; Studholme, D.J.; Moulton, V. A toolkit for analyzing large-scale plant small RNA datasets. Bioinformatics 2008, 24, 2252–2253. [Google Scholar] [CrossRef] [PubMed]
- Friedländer, M.R.; Chen, W.; Adamidi, C.; Maaskola, J.; Einspanier, R.; Knespel, S.; Rajewsky, N. Discovering microRNAs from deep sequencing data using miRDeep. Nat. Biotechnol. 2008, 26, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Friedländer, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, B.M.; Heimberg, A.M.; Moy, V.N.; Sperling, E.A.; Holstein, T.W.; Heber, S.; Peterson, K.J. The deep evolution of metazoan microRNAs. Evol. Dev. 2009, 11, 50–68. [Google Scholar] [CrossRef] [PubMed]
- Mapleson, D.; Moxon, S.; Dalmay, T.; Moulton, V. MirPlex: A tool for identifying miRNAs in high-throughput sRNA datasets without a genome. J. Exp. Zool. Part B Mol. Dev. Evol. 2013, 320, 47–56. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, X.; Kang, L. A k-mer scheme to predict piRNAs and characterize locust piRNAs. Bioinformatics 2011, 27, 771–776. [Google Scholar] [CrossRef] [PubMed]
- Menor, M.; Baek, K.; Poisson, G. Multiclass relevance units machine: Benchmark evaluation and application to small ncRNA discovery. BMC Genomics 2013, 14. [Google Scholar] [CrossRef] [PubMed]
- Hall, M.A.; Smith, L.A. Feature subset selection: A correlation based filter approach. In Proceedings of Fourth International Conference on Neural Information Processing and Intelligent Information Systems, Dunedin, New Zealand, 1997; Springer: Berlin, Germany, 1997; pp. 855–858. [Google Scholar]
- Grimson, A.; Farh, K.K-H.; Johnston, W.K.; Garrett-Engele, P.; Lim, L.P.; Bartel, D.P. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol. Cell 2007, 27, 91–105. [Google Scholar]
- Aggarwal, C.C.; Hinneburg, A.; Keim, D.A. On the surprising behavior of distance metrics in high dimensional space. In Database Theory—ICDT 2001; Springer Berlin Heidelberg: Heidelberg, Germany, 2001; pp. 420–434. [Google Scholar]
- Hall, M.; Frank, E.; Holmes, G.; Pfahringer, B.; Reutemann, P.; Witten, I.H. The WEKA data mining software: An update. ACM SIGKDD Explor. Newslett. 2009, 11, 10–18. [Google Scholar] [CrossRef]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef] [PubMed]
- Vapnik, V.N. Statistical Learning Theory, 1st ed.; Wiley-Interscience: NY, USA, 1998. [Google Scholar]
- Platt, J.C. Probabilities for SV machines. In Advances in Large Margin Classifiers; Smola, A.J., Bartlett, P.L., Scholkopf, B., Schuurmans, D., Eds.; MIT Press: Cambridge, MA, USA, 2000; pp. 61–74. [Google Scholar]
- Menor, M.; Baek, K. Relevance units machine for classification. In Proceedings of the Fourth International Conference on BioMedical Engineering and Informatics, Shanghai, China, 15–17 October 2011; pp. 2281–2285.
- Chang, C.C.; Lin, C.J. LIBSVM: A library for support vector machines. ACM Trans. Intell. Syst. Technol. 2011, 2, 1–27. [Google Scholar] [CrossRef]
- Joachims, T. Making large-scale support vector machine learning practical. In Advances in Kernel Methods; Scholkopf, B., Burges, C.J.C., Smola, A.J., Eds.; MIT Press: Cambridge, MA, USA, 1999; pp. 169–184. [Google Scholar]
- Ribeiro, B. On the evaluation of Minkovsky kernel for SVMs. Neural Parallel Sci. Comput. 2005, 13, 77–90. [Google Scholar]
- Kozomara, A.; Griffiths-Jones, S. miRBase: Intergrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011, 39, D152–D157. [Google Scholar]
- Bu, D.; Yu, K.; Sun, S.; Xie, C.; Skogerbø, G.; Miao, R.; Xiao, H.; Liao, Q.; Luo, H.; Zhao, G.; et al. NONCODE v3.0: Integrative annotation of long noncoding RNAs. Nucleic Acids Res. 2012, 40, D210–D215. [Google Scholar]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2013, 41, 991–995. [Google Scholar]
- Li, W.; Godzik, A. CD-HIT: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [PubMed]
- Kertesz, M.; Iovino, N.; Unnerstall, U.; Gaul, U.; Segal, E. The role of site accessibility in microRNA target recognition. Nat. Genet. 2007, 39, 1278–1284. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Menor, M.S.; Baek, K.; Poisson, G. Prediction of Mature MicroRNA and Piwi-Interacting RNA without a Genome Reference or Precursors. Int. J. Mol. Sci. 2015, 16, 1466-1481. https://doi.org/10.3390/ijms16011466
Menor MS, Baek K, Poisson G. Prediction of Mature MicroRNA and Piwi-Interacting RNA without a Genome Reference or Precursors. International Journal of Molecular Sciences. 2015; 16(1):1466-1481. https://doi.org/10.3390/ijms16011466
Chicago/Turabian StyleMenor, Mark S., Kyungim Baek, and Guylaine Poisson. 2015. "Prediction of Mature MicroRNA and Piwi-Interacting RNA without a Genome Reference or Precursors" International Journal of Molecular Sciences 16, no. 1: 1466-1481. https://doi.org/10.3390/ijms16011466





