Chrysin, Apigenin and Acacetin Inhibit Tumor Necrosis Factor-Related Apoptosis—Inducing Ligand Receptor-1 (TRAIL-R1) on Activated RAW264.7 Macrophages
Abstract
:1. Introduction
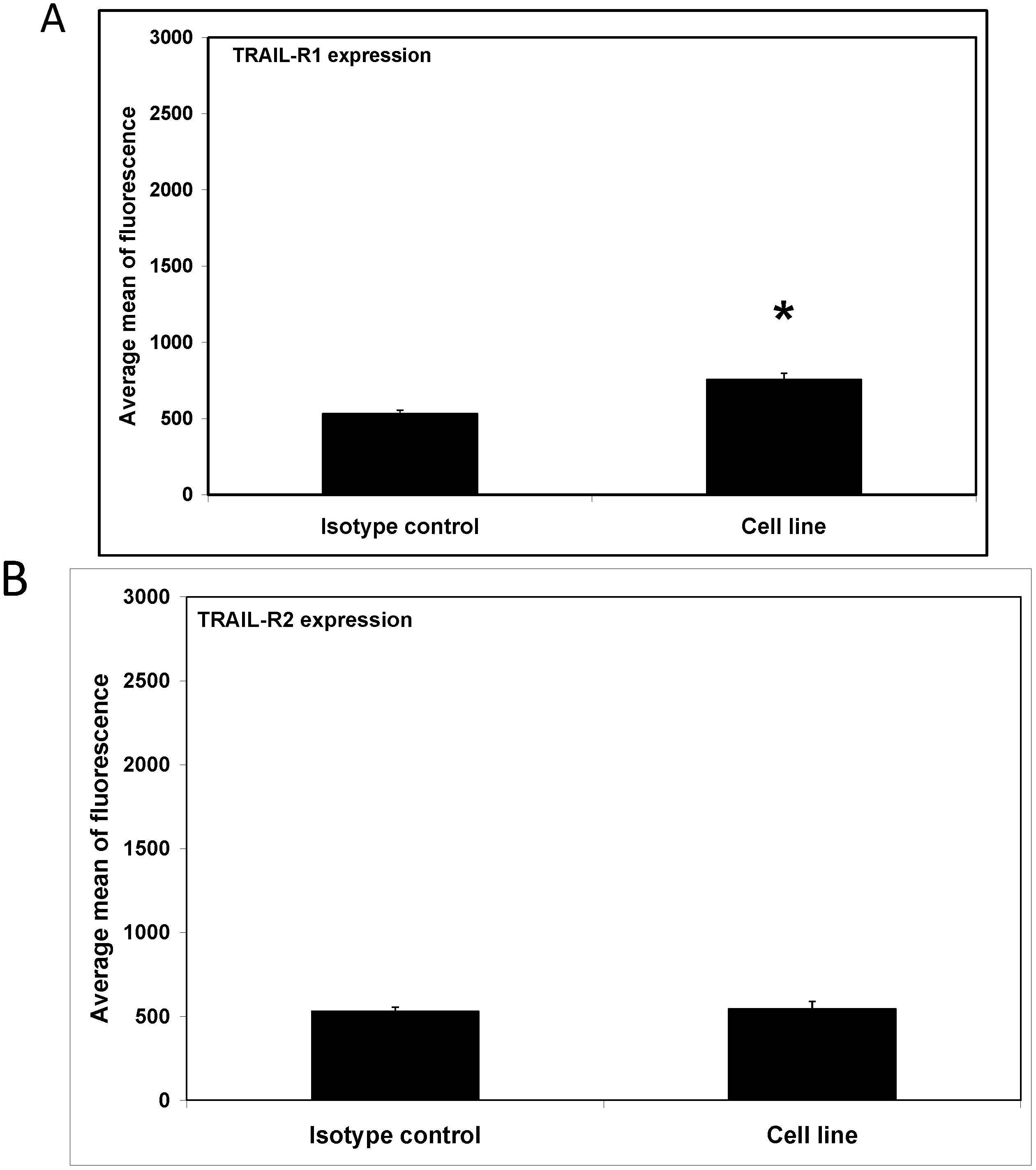
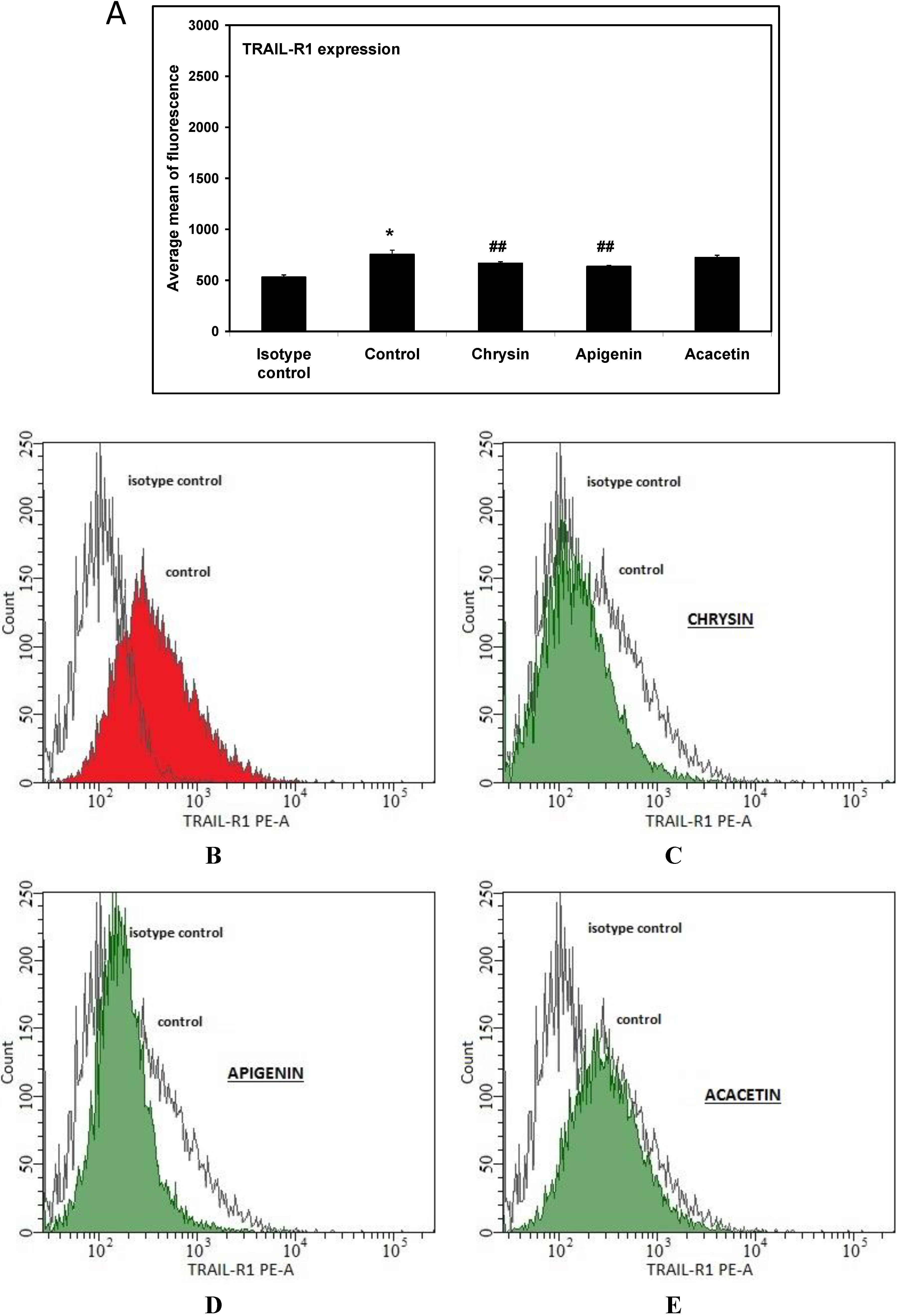
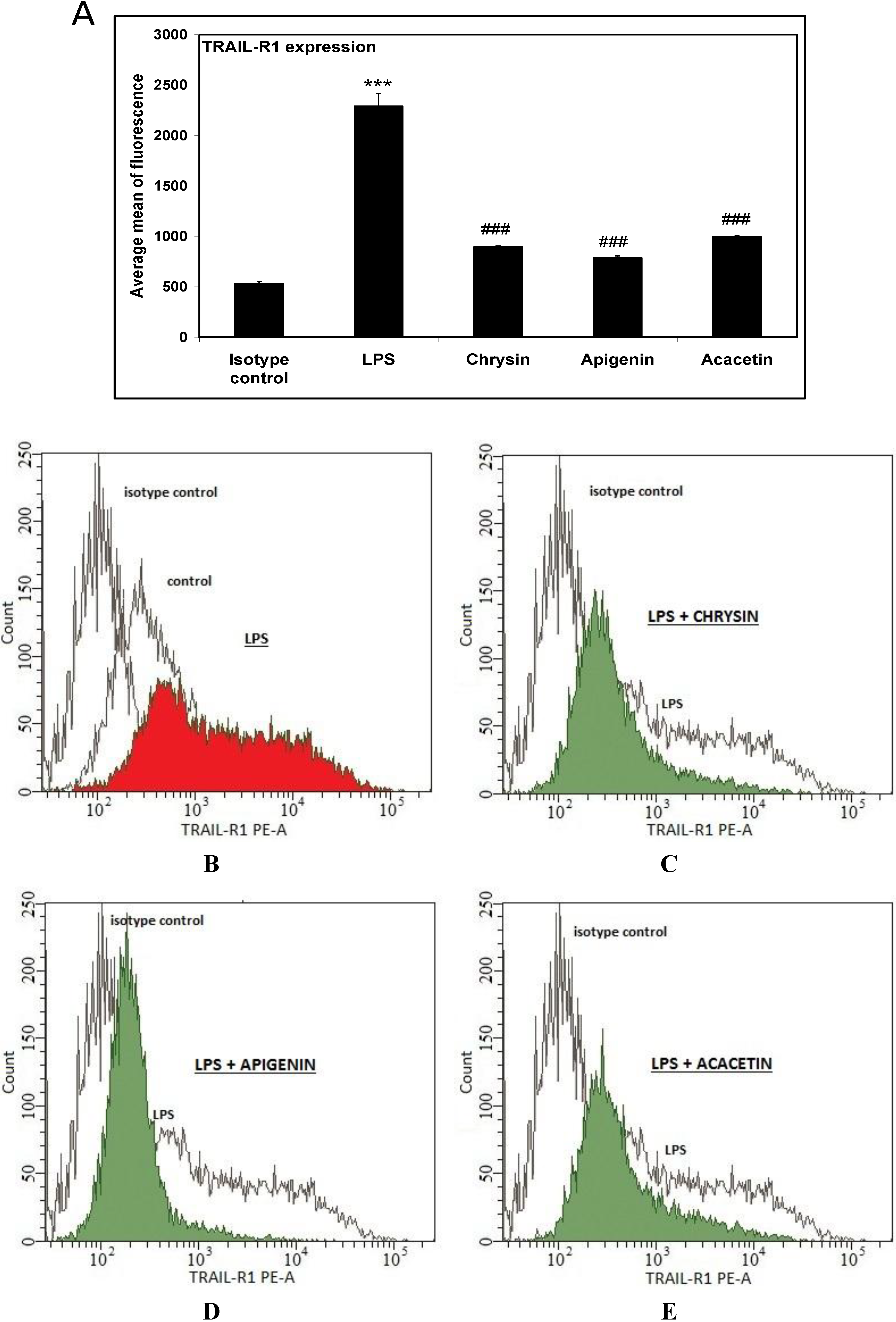
2. Results and Discussion


3. Experimental Section
3.1. Cell Culture
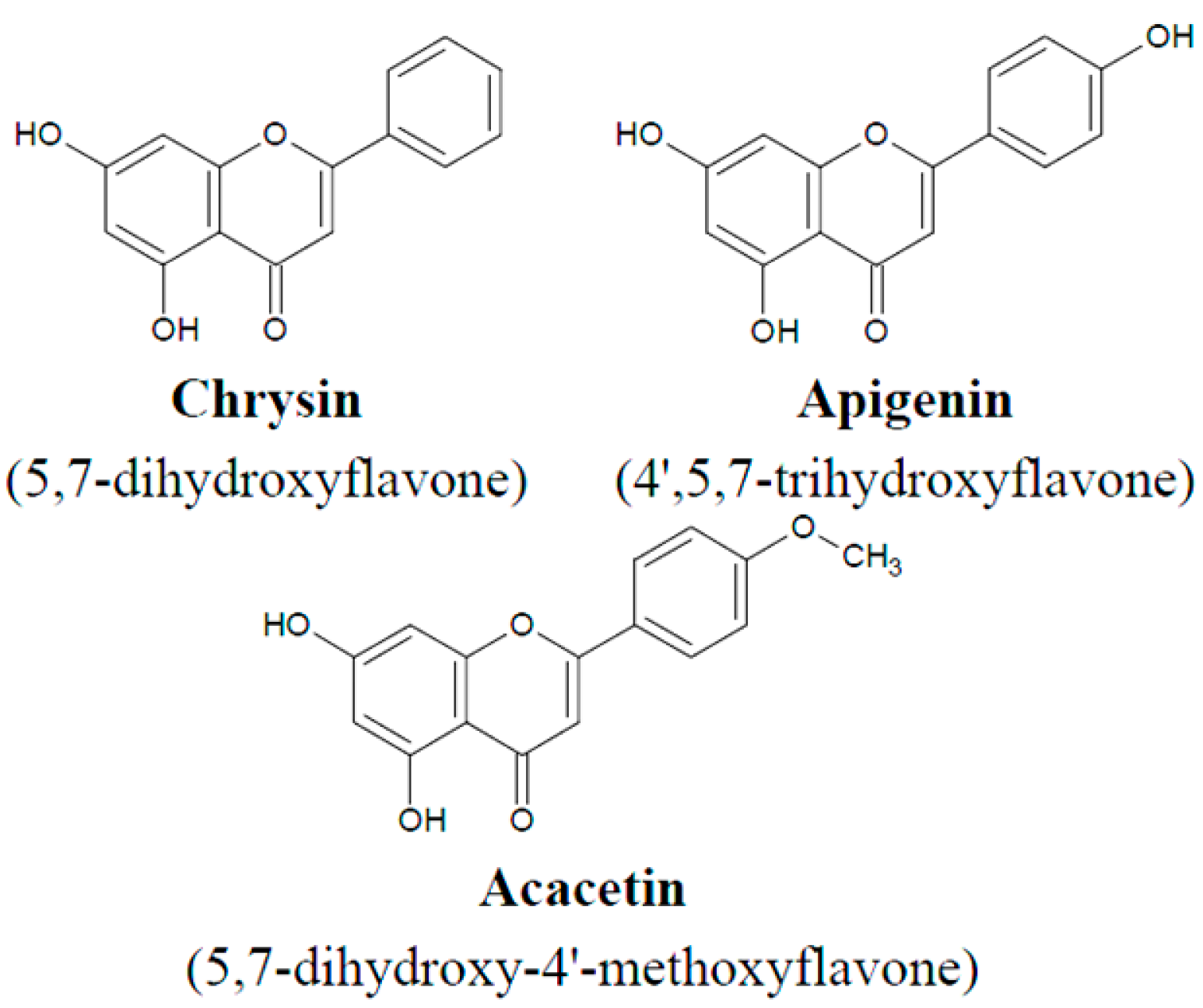
3.2. Flavones
3.3. Cell Viability
3.4. Lactate Dehydrogenase Release Assay
3.5. Flow Cytometric Analysis of Death Receptor Expression on the RAW264.7 Cells
3.6. Detection of Apoptosis by Flow Cytometry
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Willey, S.R.; Schooley, K.; Smolak, P.J.; Din, W.S.; Huang, C.P.; Nicholl, J.K.; Sutherland, G.R.; Smith, T.D.; Rauch, C.; Smith, C.A.; et al. Identification and characterization of a new member of the TNF family that induces apoptosis. Immunity 1995, 3, 673–682. [Google Scholar] [CrossRef]
- Pitti, R.M.; Marsters, S.A.; Ruppert, S.; Donahue, C.J.; Moore, A.; Ashkenazi, A. Induction of apoptosis by Apo-2 ligand, a new member of the tumor necrosis factor cytokine family. J. Biol. Chem. 1996, 271, 12687–12690. [Google Scholar]
- Fanger, N.A.; Maliszewski, C.R.; Schooley, K.; Griffith, T.S. Human dendritic cells mediate cellular apoptosis via tumor necrosis factor—Related apoptosis—Inducing ligand (TRAIL). J. Exp. Med. 1999, 190, 1155–1164. [Google Scholar] [CrossRef]
- Griffith, T.S.; Willey, S.R.; Kubin, M.Z.; Sedger, L.M.; Maliszewski, C.R.; Fanger, N.A. Monocyte-mediated tumoricidal activity via tumor necrosis factor—Related cytokine, TRAIL. J. Exp. Med. 1999, 189, 1343–1354. [Google Scholar]
- Kemp, T.J.; Ludwig, A.T.; Earel, J.K.; Moore, J.M.; Vanoosten, R.L.; Moses, B.; Leidal, K.; Nauseef, W.M.; Griffith, T.S. Neutrophil stimulation with Mycobacterium bovis Calmette-Guerin (BCG) results in the release of functional soluble TRAIL/Apo-2L. Blood 2005, 106, 3474–3482. [Google Scholar] [CrossRef]
- Kemp, T.J.; Moore, J.M.; Griffith, T.S. Human B cells express functional TRAIL/Apo2 ligand after CpG-containing oligodeoxynucleotide stimulation. J. Immunol. 2004, 173, 892–899. [Google Scholar] [CrossRef]
- Cassatella, M.A.; Huber, V.; Calzetti, F.; Margotto, D.; Tamassia, N.; Peri, G.; Mantovani, A.; Rivoltini, L.; Tecchio, C. Interferon—Activated neutrophils store a TNF—Related apoptosis—Inducing ligand (TRAIL/Apo-2 ligand) intracellular pool that is readily mobilizable following exposure to proinflammatory mediators. J. Leukoc. Biol. 2006, 79, 123–132. [Google Scholar]
- Kelley, S.K.; Ashkenazi, A. Targeting death receptor in cancer with Apo2L/TRAIL. Curr. Opin. Pharmacol. 2004, 4, 333–339. [Google Scholar] [CrossRef]
- Pan, G.; O’Rourke, K.; Chinnaiyan, A.M.; Gentz, R.; Ebner, R.; Ni, J.; Dixit, V.M. The receptor for the cytotoxic ligand TRAIL. Science 1997, 276, 111–113. [Google Scholar] [CrossRef]
- Pan, G.; Ni, J.; Wei, Y.F.; Yu, G.L.; Gentz, R.; Dixit, V.M. An antagonist decoy receptor and a death domain—Containing receptor for TRAIL. Science 1997, 277, 815–817. [Google Scholar] [CrossRef]
- Sheridan, J.P.; Marsters, S.A.; Pitti, R.M.; Gurney, A.; Skubatch, M.; Baldwin, D.; Ramakrishnan, L.; Gray, C.L.; Baker, K.; Wood, W.; et al. Control of TRAIL—Induced apoptosis by a family of signaling and decoy receptors. Science 1997, 277, 818–821. [Google Scholar] [CrossRef]
- Schneider, P.; Thome, M.; Burns, K.; Bodmer, J.L.; Hofmann, K.; Kataoka, T.; Holler, N.; Tschopp, J. TRAIL receptors 1 (DR4) and 2 (DR5) signal FADD-dependent apoptosis and activate NF-κB. Immunity 1997, 7, 831–836. [Google Scholar] [CrossRef]
- MacFarlane, M.; Ahmad, M.; Srinivasula, S.M.; Fernendes-Alnemri, T.; Cohem, G.M.; Alnemri, E.S. Identification and molecular cloning of two novel receptors for the cytotoxic ligand TRAIL. J. Biol. Chem. 1997, 272, 25417–25420. [Google Scholar]
- Screaton, G.R.; Mongkolsapaya, J.; Xu, X.N.; Cowper, A.E.; McMichael, A.J.; Bell, J.L. TRICK2, a new alternatively spliced receptor that transduces the cytotoxic signal from TRAIL. Curr. Biol. 1997, 7, 693–696. [Google Scholar] [CrossRef]
- Walczak, H.; Degli-Espositi, M.A.; Johnson, R.S.; Smolak, P.J.; Waugh, J.Y.; Boiani, N.; Timour, M.S.; Gerhart, M.J.; Schooley, K.A.; Smith, C.A.; et al. TRAIL-R2: A novel apoptosis—Mediating receptor for TRAIL. EMBO J. 1997, 16, 5386–5397. [Google Scholar] [CrossRef]
- Marsters, S.A.; Sheridan, J.P.; Pitti, R.M.; Huang, A.; Skubatch, M.; Baldwin, D.; Yuan, J.; Gurney, A.; Goddard, A.D.; Godowski, P.; et al. A novel receptor for Apo2L/TRAIL contains truncated death domain. Curr. Biol. 1997, 7, 1003–1006. [Google Scholar] [CrossRef]
- Degli-Espositi, M.A.; Dougall, W.C.; Smolak, P.J.; Waugh, J.Y.; Smith, C.A.; Goodwin, R.G. The novel receptor TRAIL-R4 induces NF-κB and protects against TRAIL-mediated apoptosis, yet retains an incomplete death domain. Immunity 1997, 7, 813–820. [Google Scholar] [CrossRef]
- Degli-Espositi, M.A.; Smolak, P.J.; Walczak, H.; Waugh, J.; Huang, C.-P.; DuBose, R.F.; Goodwin, R.G.; Smith, C.A. Cloning and characterization of TRAIL-R3, a novel member of the emerging TRAIL receptor family. J. Exp. Med. 1997, 186, 1165–1170. [Google Scholar] [CrossRef]
- Gordon, S. Macrophages and the immune system. In Fundamental Immunology, 4th ed.; Paul, W.E., Ed.; Lippincott-Raven Publishers: Philadelphia, PA, USA, 1999; pp. 533–545. [Google Scholar]
- Gordon, S.; Martinez, F.O. Alternative activation of macrophages: Mechanism and functions. Immunity 2010, 5, 593–604. [Google Scholar] [CrossRef]
- Hang, X.; Mosser, D.M. Macrophage activation by endogenous danger signals. J. Pathol. 2008, 214, 161–178. [Google Scholar]
- Mosser, D.M. The many faces of macrophage activation. J. Leukoc. Biol. 2003, 73, 209–212. [Google Scholar] [CrossRef]
- Cook, N.C.; Samman, S. Flavonoids—Chemistry, metabolism, cardioprotective effects, and dietary sources. Nutr. Biochem. 1996, 7, 66–76. [Google Scholar] [CrossRef]
- Peterson, J.; Dwyer, J. Flavonoids: Dietary occurrence and biochemical activity. Nutr. Res. 1998, 12, 1995–2018. [Google Scholar] [CrossRef]
- Patel, D.; Shukla, S.; Gupta, S. Apigenin and cancer chemoprevention: Progress, potential and promise (review). Int. J. Oncol. 2007, 30, 233–245. [Google Scholar]
- Szliszka, E.; Król, W. The role of dietary polyphenols in tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis for cancer prevention. Eur. J. Cancer Prev. 2011, 20, 63–69. [Google Scholar] [CrossRef]
- Wu, G.S.; Burns, T.F.; Zhan, Y.; Alnemri, E.S.; El-Deiry, W.S. Molecular cloning and functional analysis of the mouse homologue of the KILLER/DR5 tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) death receptor. Cancer Res. 1999, 59, 2770–2775. [Google Scholar]
- Schneider, P.; Olson, D.; Tradivel, A.; Browning, B.; Luqovskoy, A.; Gong, D.; Dobles, M.; Hertiq, S.; Hofmann, K.; van Vlijmen, H.; et al. Identification of the new murine tumor necrosis factor locus that contains two novel murine receptor for tumor necrosis factor-related apoptosis-inducing ligand (TRAIL). J. Biol. Chem. 2003, 278, 5444–5454. [Google Scholar] [CrossRef]
- Horinaka, M.; Yoshida, T.; Shiraishi, T.; Nakata, S.; Wakada, M.; Sakai, T. The dietary flavonoid apigenin sensitizes malignant tumor cells to tumor necrosis factor-related apoptosis-inducing ligand. Mol. Cancer Ther. 2006, 5, 945–951. [Google Scholar]
- Horinaka, M.; Yoshida, T.; Shiraishi, T.; Nakata, S.; Wakada, M.; Nakanishi, R.; Nishino, K.; Matsui, H.; Sakai, T. Luteolin induces apoptosis via death receptor upregulation 5 in human malignant tumor cells. Oncogene 2005, 24, 7180–7189. [Google Scholar] [CrossRef]
- Oishi, M.; Lizumi, Y.; Taniquchi, T.; Goi, W.; Miki, T.; Sakai, T. Apigenin sensitizes prostate cancer cells to Apo2L/TRAIL by targeting adenine nucleotide translocase-2. PLoS One 2013, 8, e55922. [Google Scholar]
- Kim, E.Y.; Yu, J.S.; Yang, M.; Kim, A.K. Sub-toxic dose of apigenin sensitizes HepG2 cells to TRAIL through ERK-dependent up-regulation of TRAIL receptor DR5. Mol. Cells 2013, 31, 32–40. [Google Scholar]
- Son, Y.G.; Kim, E.H.; Kim, J.Y.; Kim, S.U.; Kwon, T.K.; Yoon, A.R.; Yun, C.O.; Choi, K.S. Silibinin sensitizes human glioma cells to TRAIL-mediated apoptosis via DR5 upregulation and downregulation of cFLIP and surviving. Cancer Res. 2007, 67, 8274–8284. [Google Scholar] [CrossRef]
- Taniquchi, H.; Yoshida, T.; Horinaka, M.; Yasuda, T.; Goda, A.E.; Konishi, A.M.; Wakada, M.; Kataoka, K.; Yoshikawa, T.; Sakai, T. Baicalein overcomes tumor necrosis factor apoptosis-inducing ligand resistance via two deifferent cell-specific pathways in cancer cells but not in normal cells. Cancer Res. 2008, 68, 8918–8927. [Google Scholar] [CrossRef]
- Szliszka, E.; Mazur, B.; Żydowicz, G.; Czuba, Z.P.; Król, W. TRAIL-induced apoptosis and expression of TRAIL-R1 and TRAIL-R2 in bladder cancer cells. Folia Histochem. Cytobiol. 2009, 531, 579–585. [Google Scholar]
- Szliszka, E.; Czuba, Z.P.; Bronikowska, J.; Mertas, A.; Paradysz, A.; Król, W. Ethanolic extract of propolis (EEP) augments TRAIL-induced apoptosis death in prostate cancer cells. Evid. Based Complement. Alternat. Med. 2011. [Google Scholar] [CrossRef]
- Szliszka, E.; Skaba, D.; Czuba, Z.P.; Król, W. Inhibition of inflammatory mediators by neobavaisoflavone in activated RAW264.7 macrophages. Molecules 2011, 16, 3701–3712. [Google Scholar] [CrossRef]
- ATTC: The Global Bioresouce Center. Available online: www.lgcstandards-atcc.org/ (accessed on 19 June 2014).
- Szliszka, E.; Bronikowska, J.; Majcher, A.; Miszkiewicz, J.; Król, W. Enhanced sensitivity of hormone—refractory prostate cancer cells to tumor necrosis factor—related apoptosis—inducing ligand (TRAIL) mediated cytotoxity by taxanes. CEJ Urol. 2009, 62, 29–34. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Warat, M.; Szliszka, E.; Korzonek-Szlacheta, I.; Król, W.; Czuba, Z.P. Chrysin, Apigenin and Acacetin Inhibit Tumor Necrosis Factor-Related Apoptosis—Inducing Ligand Receptor-1 (TRAIL-R1) on Activated RAW264.7 Macrophages. Int. J. Mol. Sci. 2014, 15, 11510-11522. https://doi.org/10.3390/ijms150711510
Warat M, Szliszka E, Korzonek-Szlacheta I, Król W, Czuba ZP. Chrysin, Apigenin and Acacetin Inhibit Tumor Necrosis Factor-Related Apoptosis—Inducing Ligand Receptor-1 (TRAIL-R1) on Activated RAW264.7 Macrophages. International Journal of Molecular Sciences. 2014; 15(7):11510-11522. https://doi.org/10.3390/ijms150711510
Chicago/Turabian StyleWarat, Monika, Ewelina Szliszka, Ilona Korzonek-Szlacheta, Wojciech Król, and Zenon P. Czuba. 2014. "Chrysin, Apigenin and Acacetin Inhibit Tumor Necrosis Factor-Related Apoptosis—Inducing Ligand Receptor-1 (TRAIL-R1) on Activated RAW264.7 Macrophages" International Journal of Molecular Sciences 15, no. 7: 11510-11522. https://doi.org/10.3390/ijms150711510



