Functional Annotation of Proteomic Data from Chicken Heterophils and Macrophages Induced by Carbon Nanotube Exposure
Abstract
:1. Introduction
2. Results and Discussion
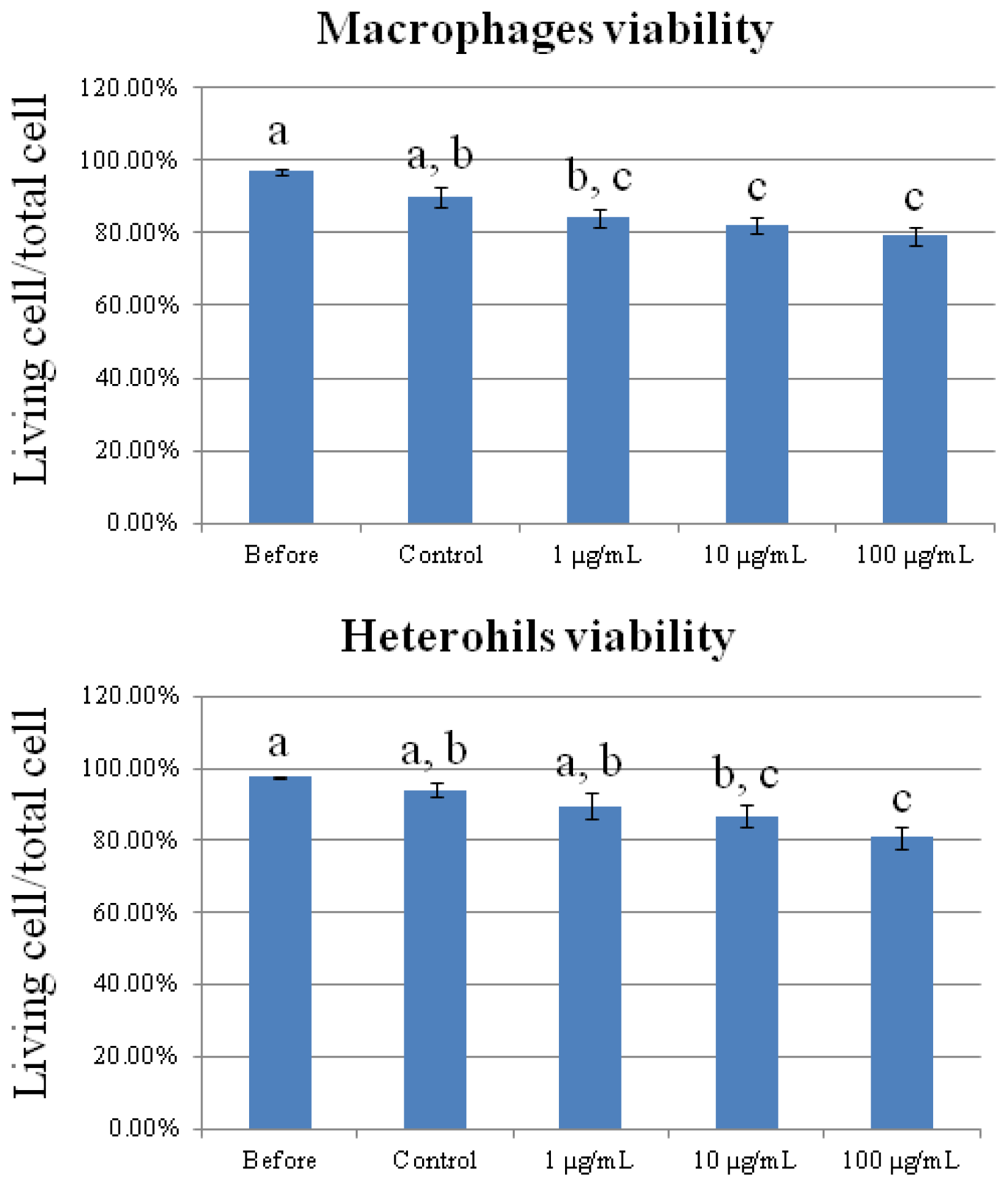
2.1. Cytotoxicity of Carbon Nanotubes on Chicken Macrophages and Heterophils
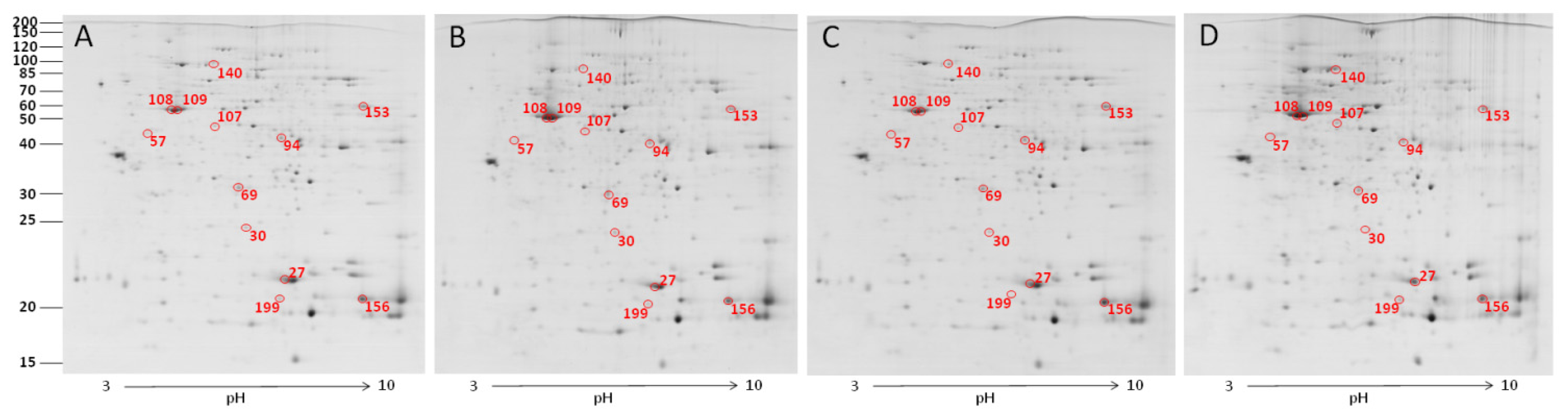
2.2. The Effect of Carbon Nanotubes on Macrophage Protein Expression
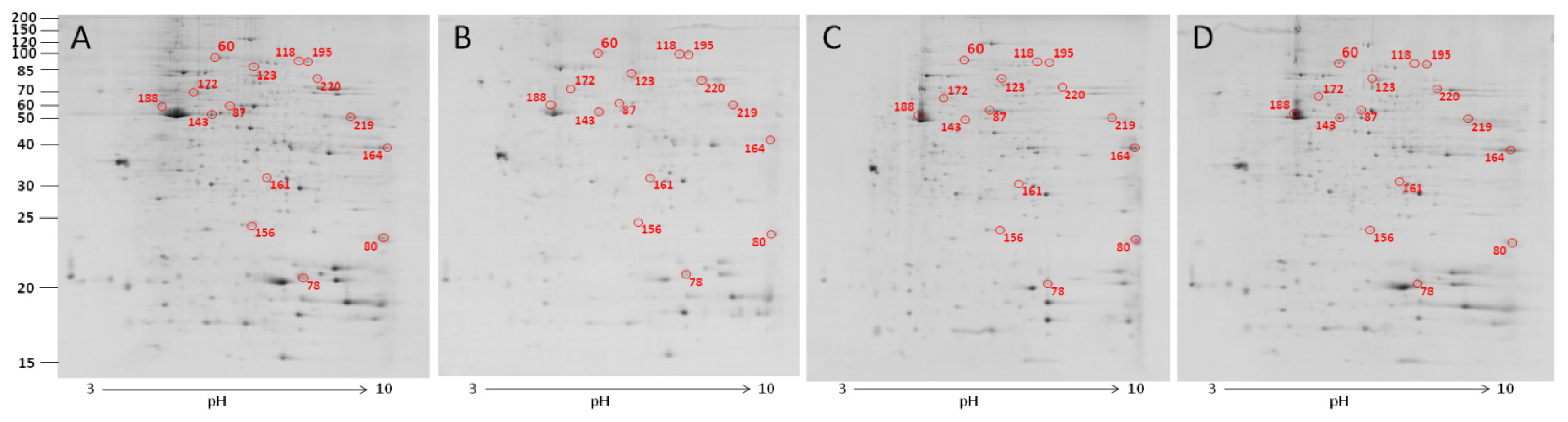
2.3. The Effect of Carbon Nanotubes on the Protein Expression of Heterophils
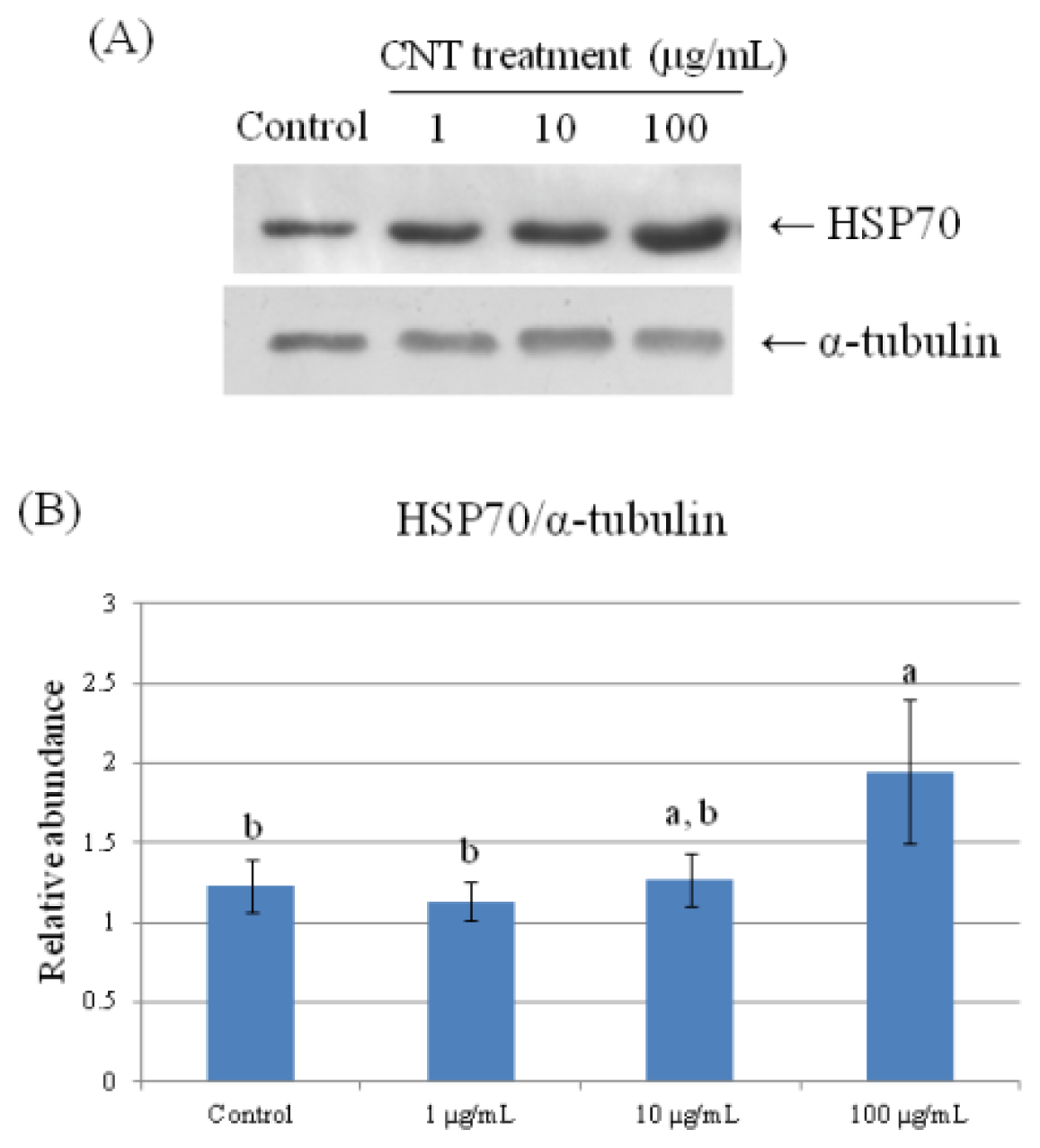
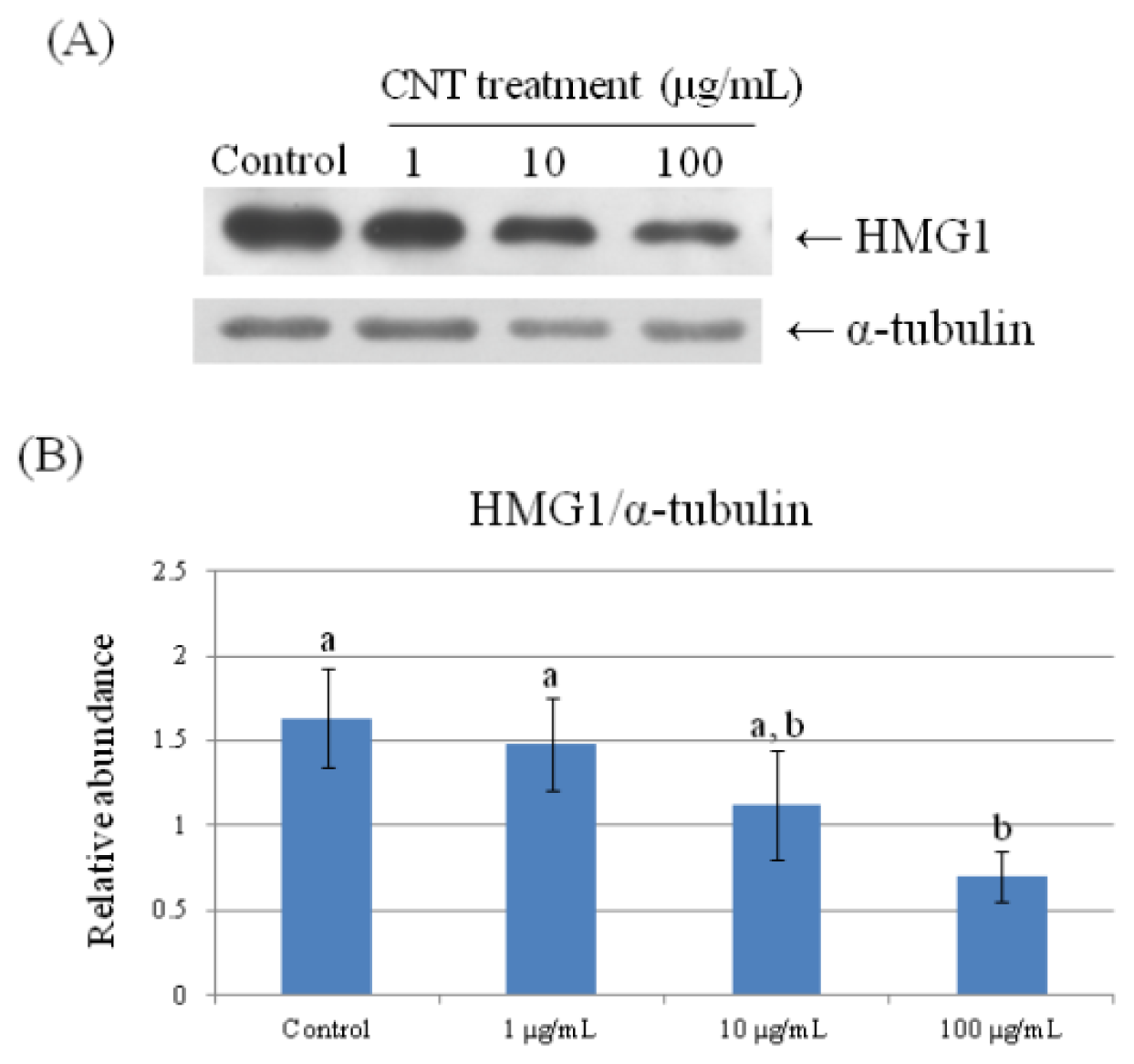
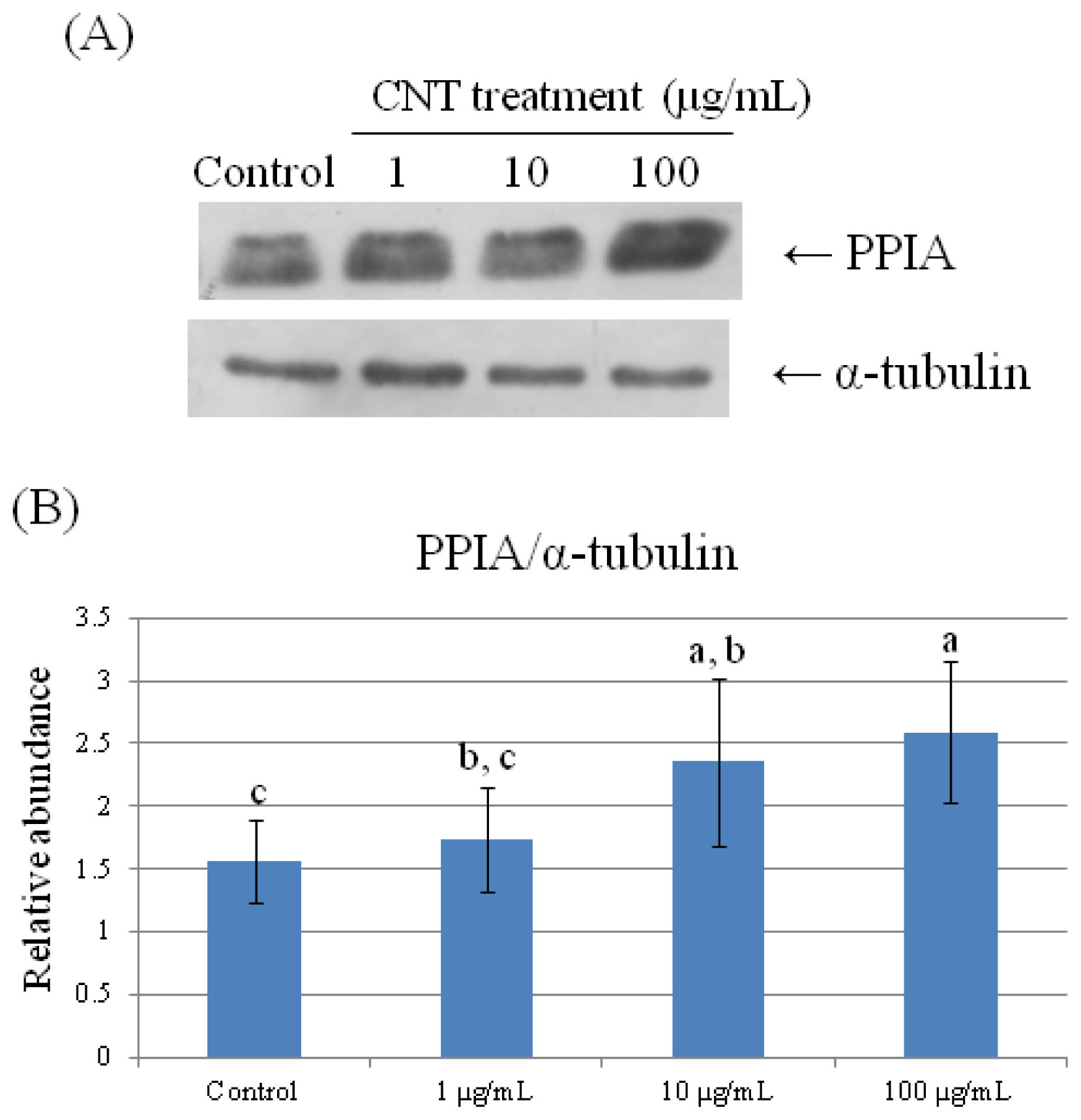
2.4. Validation of Protein Expression by Western Blot
2.5. Cell Migration Assay
2.6. IL-1β Secretion
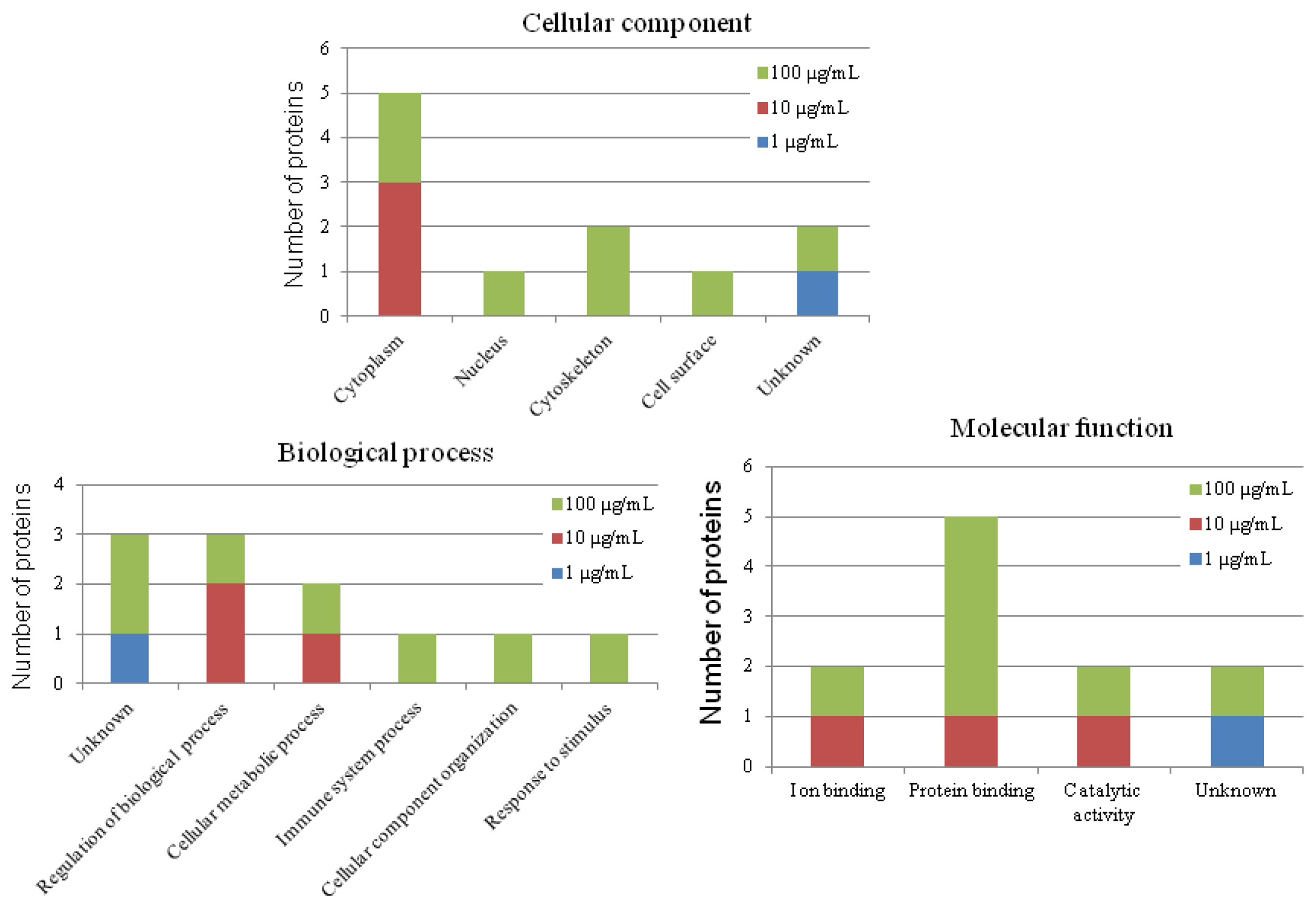
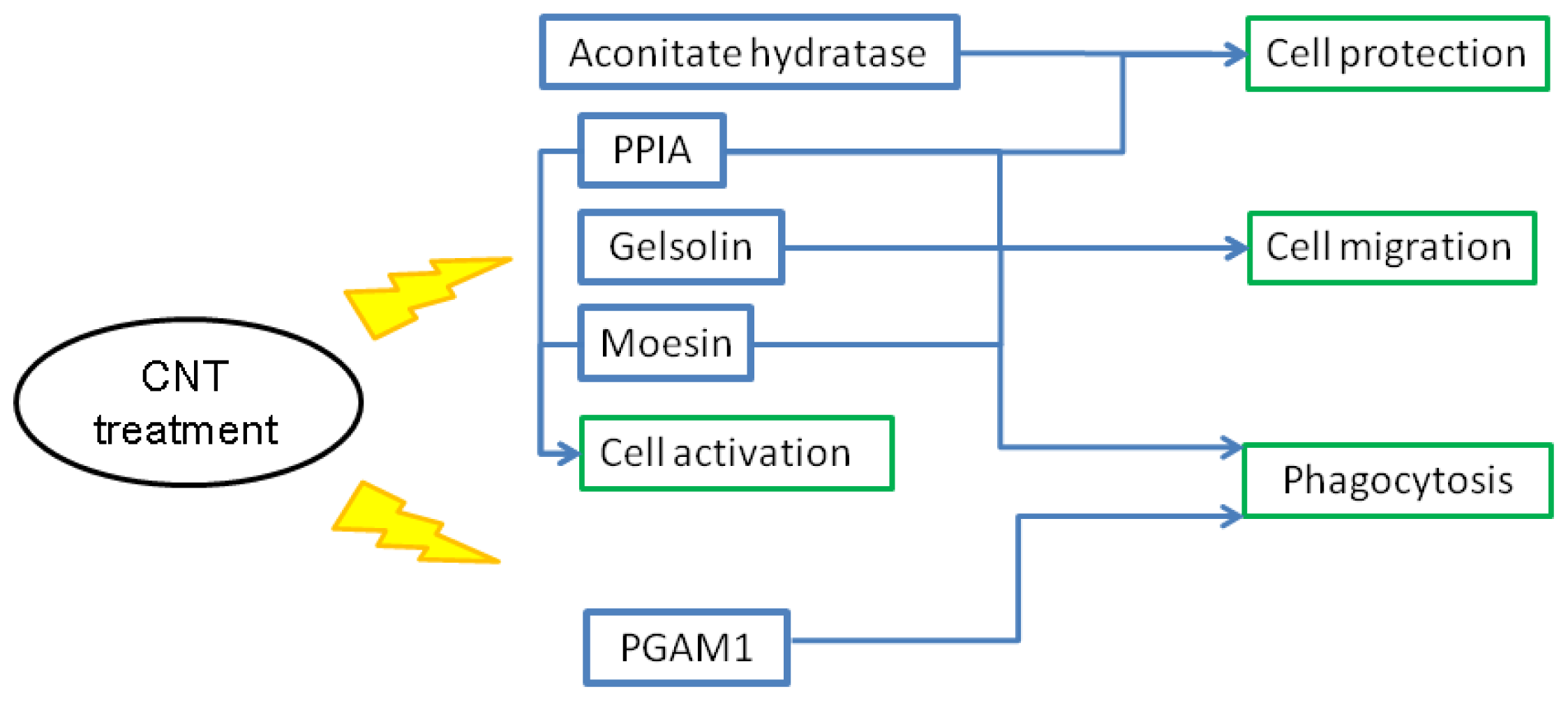
2.7. Putative Roles of the Differentially Expressed Proteins in the Functions of Chicken Macrophages and Heterophils
2.8. Possible Mechanisms of CNT-Induced Cell Death and Migration in Chicken Macrophages and Heterophils
3. Experimental Section
3.1. Source of Carbon Nanotubes
3.2. Isolation and Culture of Chicken Peripheral Blood Macrophages and Heterophils
3.3. Cell Viability Assay
3.4. Sample Preparation for Protein Analysis
3.5. Analysis of Protein by Two-Dimensional Gel Electrophoresis (2-DE)
3.6. Staining and Imaging of the 2-DE Gels
3.7. Analysis of the Variation in Protein Expression
3.8. Protein Identification by Matrix Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS) and MALDI-TOF/TOF MS
3.9. Bioinformatic Analysis of Identified Proteins
3.10. Induction of IL-1β
3.11. Western Blot Analysis
3.12. Cell Migration Assay
3.13. Statistical Analysis
4. Conclusions
Supplementary Information
ijms-15-08372-s001.pdfAcknowledgments
Conflicts of Interest
- Author ContributionsS.-Y.H. and S.-E.C. made substantial contributions to conception and design, data analysis and interpretation, and co-wrote the manuscript. Y.-Z.L. involved in collecting samples, data analysis and interpretation, and drafting the manuscript. C.-J.C. participated in data analysis and manuscript revising. C.-S.C. and Z.-L.L. took part in collecting samples, data analysis and interpretation, and manuscript revising. Y.-T.L. participated in the coordination of the study and revised critically the manuscript for important intellectual content. All authors have read and approved the final manuscript to be submitted.
References
- Curtis, J.; Greenberg, M.; Kester, J.; Phillips, S.; Krieger, G. Nanotechnology and nanotoxicology: A primer for clinicians. Toxicol. Sci 2006, 25, 245–260. [Google Scholar]
- Stern, S.T.; McNeil, S.E. Nanotechnology safety concerns revisited. Toxicol. Sci 2008, 101, 4–21. [Google Scholar]
- Deng, X.; Xiong, D.; Wang, Y.; Chen, W.; Luan, Q.; Zhang, H.; Jiao, Z.; Wu, M. Water soluble multi-walled carbon nanotubes enhance peritoneal macrophage activity in vivo. J. Nanosci. Nanotechnol. 2010, 12, 8663–8669. [Google Scholar]
- Li, J.; Yang, F.; Guo, G.; Yang, D.; Long, J.D.F. Preparation of biocompatible multiwalled carbon nanotubes as potential tracers for sentinel lymph nodes. Polym. Int 2010, 59, 169–174. [Google Scholar]
- Sinha, R.; Kim, G.J.; Nie, S.; Shin, D.M. Nanotechnology in cancer therapeutics: Bioconjugated nanoparticles for drug delivery. Mol. Cancer Ther 2006, 5, 1909–1917. [Google Scholar]
- Wang, X.; Jia, G.; Wang, H.; Nie, H.; Yan, L.; Deng, X.Y.; Wang, S. Diameter effects on cytotoxicity of multi-walled carbon nanotubes. J. Nanosci. Nanotechnol 2009, 5, 3025–3033. [Google Scholar]
- Maynard, A.D.; Baron, P.A.; Foley, M.; Shvedova, A.A.; Kisin, E.R.; Castranova, V. Exposure to carbon nanotube material: Aerosol release during the handling of unrefined single-walled carbon nanotube material. J. Toxicol. Environ. Health 2004, 67, 87–107. [Google Scholar]
- Lam, C.W.; James, J.T.; McCluskey, R.; Hunter, R.L. Pulmonary toxicity of single-wall carbon nanotubes in mice 7 and 90 days after intratracheal instillation. Toxicol. Sci 2004, 77, 126–134. [Google Scholar]
- Shvedova, A.A.; Kisin, E.R.; Mercer, R.; Murray, A.R.; Johnson, V.J.; Potapovich, A.I.; Tyurina, Y.Y.; Gorelik, O.; Arepalli, S.; Schwegler-Berry, D.; et al. Unusual inflammatory and fibrogenic pulmonary responses to singlewalled carbon nanotubes in mice. Am. J. Physiol. Lung Cell Mol. Physiol 2005, 289, 698–708. [Google Scholar]
- Warheit, D.B.; Laurence, B.R.; Reed, K.L.; Roach, D.H.; Reynolds, G.A.; Webb, T.R. Comparative pulmonary toxicity assessment of single-wall carbon nanotubes in rats. Toxicol. Sci 2004, 77, 117–125. [Google Scholar]
- Muller, J.; Decordier, I.; Hoet, P.; Lombaert, N.; Thomassen, L.; Huaux, F.; Lison, D.; Kirsch-Volders, M. Clastogenic and aneugenic effects of multi-wall carbon nanotubes in epithelial cells. Carcinogenesis 2008, 29, 427–433. [Google Scholar]
- Bottini, M.; Bruckner, S.; Nika, K.; Bottini, N.; Bellucci, S.; Magrini, A.; Bergamaschi, A.; Mustelin, T. Multi-walled carbon nanotubes induce T lymphocyte apoptosis. Toxicol. Lett 2006, 160, 121–126. [Google Scholar]
- Cui, D.; Tian, F.; Ozkan, C.S.; Wang, M.; Gao, H. Effect of single-wall carbon nanotubes on human HEK293 cells. Toxicol. Lett 2005, 155, 73–85. [Google Scholar]
- Monteiro-Riviere, N.A.; Nemanich, R.J.; Inman, A.O.; Wang, Y.Y.; Riviere, J.E. Multi-walled carbon nanotube interactions with human epidermal keratinocytes. Toxicol. Lett 2005, 155, 377–384. [Google Scholar]
- Deng, X.; Jia, G.; Sun, H.F.; Wang, H.F.; Wang, X.; Yang, S.T.; Wang, T.C.; Liu, Y.F. Translocation and fate of multi-walled carbon nanotubes in vivo. Carbon 2007, 45, 1419–1424. [Google Scholar]
- Jia, G.; Wang, H.; Yan, L.; Wang, X.; Pei, R.; Yan, T.; Zhao, Y.; Guo, X. Cytotoxicity of carbon nanomaterials: Single-wall nanotube, multi-wall nanotube, and fullerene. Environ. Sci. Technol 2005, 39, 1378–1383. [Google Scholar]
- Maxwell, M.H.; Robertson, G.W. The avian heterophil leucocyte: A review. World’s Poult. Sci. J 1998, 54, 155–178. [Google Scholar]
- Sheehan, D. The potential of proteomics for providing new insights into environmental impacts on human health. Rev. Environ. Health 2007, 22, 175–194. [Google Scholar]
- Tian, F.; Cui, D.; Sehwarz, H.; Estrada, G.G.; Kobayashi, H. Cytotoxicity of single-wall carbon nanotube on human fibroblasts. Toxicol. in Vitro 2006, 20, 1202–1212. [Google Scholar]
- Manna, S.K.; Sarkar, S.; Barr, J.; Wise, K.; Barrera, E.V.; Jejelowo, O.; Rice-Ficht, A.C.; Ramesh, G.T. Single-walled carbon nanotubes induces oxidative stress and activates nuclear transcription factor-kappaB in human keratinocytes. Nano Lett 2005, 5, 1676–1684. [Google Scholar]
- Ye, S.F.; Wu, Y.H.; Hou, Z.Q.; Zhang, Q.Q. ROS and NF-kappaB are involved in upregulation of IL-8 in A549 cells exposed to multi-walled carbon nanotubes. Biochem. Biophys. Res. Commun 2009, 379, 643–648. [Google Scholar]
- Pinkus, R.; Weiner, L.M.; Daniel, V. Role of oxidants and antioxidants in the induction of AP-1, NF-κB, and glutathione S-transferase gene expression. J. Biol. Chem 1996, 271, 13422–13429. [Google Scholar]
- Shvedova, A.A.; Fabisiak, J.P.; Kisi, E.R.; Murra, A.R.; Roberts, J.R.; Tyurina, Y.Y.; Antonini, J.M.; Feng, W.H.; Kommineni, C.; Reynolds, J.; et al. Sequential exposure to carbon nanotubes and bacteria enhances pulmonary inflammation and infectivity. Am. J. Respir. Cell Mol. Biol 2008, 38, 579–590. [Google Scholar]
- Schrand, A.M.; Dai, L.; Schlager, J.J.; Hussain, S.M.; Osawa, E. Differential bio compatibly of carbon nanotubes and nanodiamonds. Diamond Relat. Mater 2007, 16, 2118–2123. [Google Scholar]
- He, X.; Young, S.H.; Schwegler-Berry, D.; Chisholm, W.P.; Fernback, J.E.; Ma, Q. Multiwalled carbon nanotubes induce a fibrogenic response by stimulating reactive oxygen species production, activating NF-κB signaling, and promoting fibroblast-to-myofibroblast transformation. Chem. Res. Toxicol 2011, 24, 2237–2248. [Google Scholar]
- Yadav, R.; Larbi, K.Y.; Young, R.E.; Nourshargh, S. Migration of leukocytes through the vessel wall and beyond. Thromb. Haemost 2003, 90, 598–606. [Google Scholar]
- Tavaria, M.; Gabriele, T.; Kola, I.; Anderson, R.L. A hitchhiker’s guide to the human Hsp70 family. Cell Stress Chaperones 1996, 1, 3–28. [Google Scholar]
- Basu, S.; Binder, R.J.; Suto, R.; Anderson, K.M.; Srivastava, P.K. Necrotic but not apoptotic cell death releases heat shock proteins, which deliver a partial maturation signal to dendritic cells and activate the NF-κB pathway. Int. Immunol 2000, 12, 1539–1546. [Google Scholar]
- Pawaria, S.J.; Binder, R. CD91-dependent modulation of immune responses by heat shock proteins: A role in autoimmunity. Autoimmune Dis 2012, 2012. [Google Scholar] [CrossRef]
- Novakova, M.; Dolezal, T. Expression of Drosophila adenosine deaminase in immune cells during inflammatory response. PLoS One 2011, 6, e17741. [Google Scholar]
- Pérez-Aguilar, M.C.; Goncalves, L.; Ibarra, A.; Bonfante-Cabarcas, R. Adenosine deaminase as costimulatory molecule and marker of cellular immunity. Investig. Clin 2010, 51, 561–571. [Google Scholar]
- Tsan, M.F. Heat shock proteins and high mobility group box 1 protein lack cytokine function. J. Leukoc. Biol 2011, 89, 847–853. [Google Scholar]
- Park, J.S.; Gamboni-Robertson, F.; He, Q.; Svetkauskaite, D.; Kim, J.Y.; Strassheim, D.; Sohn, J.W.; Yamada, S.; Maruyama, I.; Banerjee, A.; et al. High mobility group box 1 protein interacts with multiple Toll-like receptors. Am. J. Physiol. Cell Physiol 2006, 290, 917–924. [Google Scholar]
- Wang, H.; Bloom, O.; Zhang, M.; Vishnubhakat, J.M.; Ombrellino, M.; Che, J.; Frazier, A.; Yang, H.; Ivanova, S.; Borovikova, L.; et al. HMG-1 as a late mediator of endotoxin lethality in mice. Science 1999, 285, 248–251. [Google Scholar]
- Feng, L.; Zhu, M.; Zhang, M.; Jia, X.; Cheng, X.; Ding, S.; Zhu, Q. Amelioration of compound 4,4′-diphenylmethane-bis(methyl)carbamate on high mobility group box1-mediated inflammation and oxidant stress responses in human umbilical vein endothelial cells via RAGE/ERK1/2/NF-κB pathway. Int. Immunopharmacol 2013, 15, 206–216. [Google Scholar]
- Dugina, V.; Zwaenepoel, I.; Gabbiani, G.; Clement, S.; Chaponnier, C. β and γ-cytoplasmic actins display distinct distribution and functional diversity. J. Cell Sci 2009, 122, 2980–2988. [Google Scholar]
- Vallenius, T.; Mäkelä, T.P. Clik1: A novel kinase targeted to actin stress fibers by the CLP-36 PDZ-LIM protein. J. Cell Sci 2002, 115, 2067–2073. [Google Scholar]
- Patel, D.; Ahmad, S.; Weiss, D.; Gerke, V.; Kuznetsov, S. Annexin A1 is a new functional linker between actin filaments and phagosomes during phagocytosis. J. Cell Sci 2011, 124, 578–588. [Google Scholar]
- Tanaka, H.; Shirkoohi, R.; Nakagawa, K.; Qiao, H.; Fujita, H.; Okada, F.; Hamada, J.; Kuzumaki, S.; Takimoto, M.; Kuzumaki, N. siRNA gelsolin knockdown induces epithelial-mesenchymal transition with a cadherin switch in human mammary epithelial cells. Int. J. Cancer 2006, 118, 1680–1691. [Google Scholar]
- Oikonomou, N.; Thanasopoulou, A.; Tzouvelekis, A.; Harokopos, V.; Paparountas, T.; Nikitopoulou, I.; Witke, W.; Karameris, A.; Kotanidou, A.; Bouros, D.; et al. Gelsolin expression is necessary for the development of modeled pulmonary inflammation and fibrosis. Thorax 2009, 64, 467–475. [Google Scholar]
- Arpin, M.; Chirivino, D.; Naba, A.; Zwaenepoel, I. Emerging role for ERM proteins in cell adhesion and migration. Cell Adh. Migr 2011, 5, 199–206. [Google Scholar]
- Boldt, K.; Rist, W.; Weiss, S.M.; Weith, A.; Lenter, M.C. FPRL-1 induces modifications of migration-associated proteins in human neutrophils. Proteomics 2006, 6, 4790–4799. [Google Scholar]
- Amar, S.; Oyaisu, K.; Li, L.; van Dyke, T. Moesin: A potential LPS receptor on human monocytes. J. Endotoxin Res 2001, 7, 281–286. [Google Scholar]
- Ruddon, R.W.; Bedows, E. Assisted protein folding. J. Biol. Chem 1997, 272, 3125–3128. [Google Scholar]
- Zhang, X.C.; Wang, W.D.; Wang, J.S.; Pan, J.C. PPIase independent chaperone-like function of recombinant human Cyclophilin A during arginine kinase refolding. FEBS Lett 2013, 587, 666–672. [Google Scholar]
- Sherry, B.; Yarlett, N.; Strupp, A.; Cerami, A. Identification of cyclophilin as a proinflammatory secretory product of lipopolysaccharide-activated macrophages. Proc. Natl. Acad. Sci. USA 1992, 89, 3511–3515. [Google Scholar]
- Arora, K.; Gwinn, W.M.; Bower, M.A.; Watson, A.; Okwumabua, I.; MacDonald, H.R.; Bukrinsky, M.I.; Constant, S.L. Extracellular cyclophilins contribute to the regulation of inflammatory responses. J. Immunol 2005, 175, 517–522. [Google Scholar]
- Zou, G.M.; Tam, Y.K. Cytokines in the generation and maturation of dendritic cells: recent advances. Eur. Cytokine Netw 2002, 13, 186–199. [Google Scholar]
- Jortner, B.S.; Adams, W.R. Turpentine-induced inflammation in the chicken. A light and electronmicroscope study, with emphasis on the macrophage, epithelioid cell, and multinucleated giant cell reaction. Avian Dis 1971, 15, 533–550. [Google Scholar]
- Cheng, X.L.; Zhong, J.; Meng, J.; Yang, M.; Jia, F.; Xu, Z.; Kong, H.; Xu, H.Y. Characterization of multiwalled carbon nanotubes dispersing in water and association with biological effects. J. Nanomater 2011. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem 1976, 72, 248–254. [Google Scholar]
- Görg, A.; Weissn, W.; Dunn, M.J. Current state of two-dimensional electrophoresis technology for proteomics. Proteomics 2004, 4, 3665–3685. [Google Scholar]
- Huang, S.Y.; Lin, J.H.; Chen, Y.H.; Chuang, C.K.; Lin, E.C.; Huang, M.C.; Sun, H.S.; Lee, W.C. A reference map and identification of porcine testis proteins using two-dimensional gel electrophoresis and mass spectrometry. Proteomics 2005, 5, 4205–4212. [Google Scholar]
- Neuhoff, V.; Arold, N.; Taube, D.; Ehrhardt, W. Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis 1988, 9, 255–262. [Google Scholar]
- Huang, S.Y.; Chen, Y.H.; Teng, S.H.; Chen, I.C.; Ho, L.L.; Tu, C.F. Protein expression of lymphocytes in HLA-DR transgenic pigs by a proteomic approach. Proteomics 2006, 6, 5815–5825. [Google Scholar]
- Havlis, J.; Thomas, H.; Sebela, M.; Shevchenko, A. Fast-response proteomics by accelerated in-gel digestion of proteins. Anal. Chem 2003, 75, 1300–1306. [Google Scholar]
- Perkins, D.N.; Pappin, D.J.; Creasy, D.M.; Cottrell, J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 1999, 20, 3551–3567. [Google Scholar]
- UniProt Database 2012. Available online: http://www.uniprot.org accessed on 14 March 2014.
- The Gene Ontology Database 2012. Available online: http://www.geneontology.org accessed on 14 March 2014.
- Jang, S.I.; Lillehoj, H.S.; Lee, S.H.; Kim, D.K.; Pagés, M.; Hong, Y.H.; Min, W.; Lillehoj, E.P. Distinct immunoregulatory properties of macrophage migration inhibitory factors encoded by Eimeria parasites and their chicken host. Vaccine 2011, 29, 8998–9004. [Google Scholar]
- SAS, SAS/STAT User’s Guide; Version 9.2 ed; SAS Institute Inc: Cary, NC, USA, 2010.




| Spot No. * | Protein Identity | Concentration of Carbon Nanotube | |||
|---|---|---|---|---|---|
| Control (0 μg/mL) | 1 μg/mL | 10 μg/mL | 100 μg/mL | ||
| 27 | Protein MRP-126 | 1.310 ± 0.128 b | 1.464 ± 0.082 a,b | 1.838 ± 0.363 a | 1.182 ± 0.042 b |
| 30 | similar to D4-GDP-dissociation inhibitor | 0.290 ± 0.058 a,b | 0.332 ± 0.031 a | 0.269 ± 0.013 a,b | 0.196 ± 0.049 b |
| 57 | similar to hepatoma-derived growth factor (high-mobility group protein 1-like) | 0.193 ± 0.033 b | 0.191 ± 0.027 b | 0.298 ± 0.030 a | 0.218 ± 0.027 a,b |
| 69 | high mobility group protein HMG1 | 0.660 ± 0.061 a | 0.622 ± 0.015 a | 0.530 ± 0.113 a,b | 0.351 ± 0.019 b |
| 94 | similar to Pdlim1 protein isoform 1 | 0.378 ± 0.051 a | 0.410 ± 0.049 a | 0.407 ± 0.022 a | 0.161 ± 0.056 b |
| 107 | adenosine deaminase | 0.168 ± 0.011 b | 0.161 ± 0.021 b | 0.185 ± 0.009 b | 0.254 ± 0.020 a |
| 108 | actin, cytoplasmic type 5 | 2.256 ± 0.669 b,c | 1.848 ± 0.534 c | 3.833 ± 0.837 a,b | 4.067 ± 0.269 a |
| 109 | beta-actin | 2.186 ± 0.241 a,b | 2.958 ± 0.316 a | 1.806 ± 0.248 b | 2.337 ± 0.197 a,b |
| 140 | heat shock protein 70 | 0.337 ± 0.041 b | 0.330 ± 0.027 b | 0.310 ± 0.047 b | 0.488 ± 0.056 a |
| 153 | phosphoglycerate kinase | 0.975 ± 0.117 a | 0.733 ± 0.093 a,b | 0.484 ± 0.116 b | 0.747 ± 0.152 a,b |
| 156 | NS | 3.204 ± 0.373 a,b | 2.855 ± 0.518 a,b | 1.852 ± 0.207 b | 4.010 ± 1.104 a |
| 199 | NS | 0.716 ± 0.082 a | 0.389 ± 0.069 b | 0.505 ± 0.134 a,b | 0.389 ± 0.061 b |
| Spot No. * | Protein Identity | Concentration of Carbon Nanotube | |||
|---|---|---|---|---|---|
| Control (0 μg/mL) | 1 μg/mL | 10 μg/mL | 100 μg/mL | ||
| 60 | gelsolin precursor | 0.131 ± 0.018 b | 0.168 ± 0.029 a,b | 0.201 ± 0.037 a,b | 0.227 ± 0.030 a |
| 78 | peptidylprolyl isomerase A (cyclophilin A) | 0.723 ± 0.191 b | 0.754 ± 0.090 b | 0.885 ± 0.191 a,b | 1.275 ± 0.057 a |
| 80 | recombination activating protein 1 | 0.713 ± 0.043 b | 1.191 ± 0.222 a | 1.359 ± 0.163 a | 1.660 ± 0.213 a |
| 87 | NS | 0.103 ± 0.002 b | 0.212 ± 0.028 a | 0.161 ± 0.033 a,b | 0.186 ± 0.042 a,b |
| 118 | aconitate hydratase, mitochondrial | 0.063 ± 0.016 b | 0.109 ± 0.023 a,b | 0.131 ± 0.026 a | 0.093 ± 0.010 a,b |
| 123 | moesin-like | 0.221 ± 0.008 a,b | 0.222 ± 0.009 a,b | 0.286 ± 0.058 a | 0.175 ± 0.011 b |
| 143 | moesin-like | 0.151 ± 0.039 b | 0.178 ± 0.034 a,b | 0.197 ± 0.028 a,b | 0.272 ± 0.012 a |
| 156 | cell division control protein 42 homolog precursor | 0.255 ± 0.012 a,b | 0.309 ± 0.040 a | 0.186 ± 0.017 b | 0.225 ± 0.018 b |
| 161 | phosphoglycerate mutase 1 | 0.161 ± 0.022 b | 0.203 ± 0.006 a,b | 0.169 ± 0.013 b | 0.250 ± 0.015 a |
| 164 | glyceraldehyde-3-phosphate dehydrogenase | 0.737 ± 0.143 b | 1.338 ± 0.144 a | 0.777 ± 0.219 b | 0.411 ± 0.077 b |
| 172 | peptidyl-prolyl cis-trans isomerase FKBP4 | 0.077 ± 0.007 b | 0.138 ± 0.028 a | 0.103 ± 0.003 a,b | 0.084 ± 0.015 b |
| 188 | beta-actin | 0.246 ±0.045 a,b | 0.254 ± 0.068 a,b | 0.356 ± 0.092 a | 0.150 ± 0.015 b |
| 195 | hypothetical protein RCJMB04_1a14 | 0.110 ± 0.023 a | 0.063 ± 0.005 b | 0.073 ± 0.007 a,b | 0.063 ± 0.005 b |
| 219 | phosphoglycerate kinase | 0.905 ± 0.050 a | 0.563 ± 0.131 b | 0.833 ± 0.064 a | 0.525 ± 0.092 b |
| 220 | similar to transketolase | 0.195 ± 0.013 a | 0.171 ± 0.028 a | 0.149 ± 0.011 a,b | 0.112 ± 0.011 b |
© 2014 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Li, Y.-Z.; Cheng, C.-S.; Chen, C.-J.; Li, Z.-L.; Lin, Y.-T.; Chen, S.-E.; Huang, S.-Y. Functional Annotation of Proteomic Data from Chicken Heterophils and Macrophages Induced by Carbon Nanotube Exposure. Int. J. Mol. Sci. 2014, 15, 8372-8392. https://doi.org/10.3390/ijms15058372
Li Y-Z, Cheng C-S, Chen C-J, Li Z-L, Lin Y-T, Chen S-E, Huang S-Y. Functional Annotation of Proteomic Data from Chicken Heterophils and Macrophages Induced by Carbon Nanotube Exposure. International Journal of Molecular Sciences. 2014; 15(5):8372-8392. https://doi.org/10.3390/ijms15058372
Chicago/Turabian StyleLi, Yun-Ze, Chung-Shi Cheng, Chao-Jung Chen, Zi-Lin Li, Yao-Tung Lin, Shuen-Ei Chen, and San-Yuan Huang. 2014. "Functional Annotation of Proteomic Data from Chicken Heterophils and Macrophages Induced by Carbon Nanotube Exposure" International Journal of Molecular Sciences 15, no. 5: 8372-8392. https://doi.org/10.3390/ijms15058372
APA StyleLi, Y.-Z., Cheng, C.-S., Chen, C.-J., Li, Z.-L., Lin, Y.-T., Chen, S.-E., & Huang, S.-Y. (2014). Functional Annotation of Proteomic Data from Chicken Heterophils and Macrophages Induced by Carbon Nanotube Exposure. International Journal of Molecular Sciences, 15(5), 8372-8392. https://doi.org/10.3390/ijms15058372






