RNA Sequencing Analysis Reveals Transcriptomic Variations in Tobacco (Nicotiana tabacum) Leaves Affected by Climate, Soil, and Tillage Factors
Abstract
:1. Introduction
2. Results and Discussion
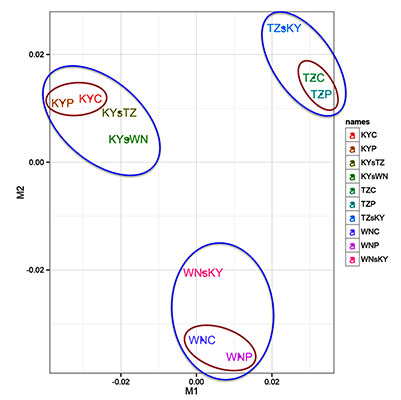
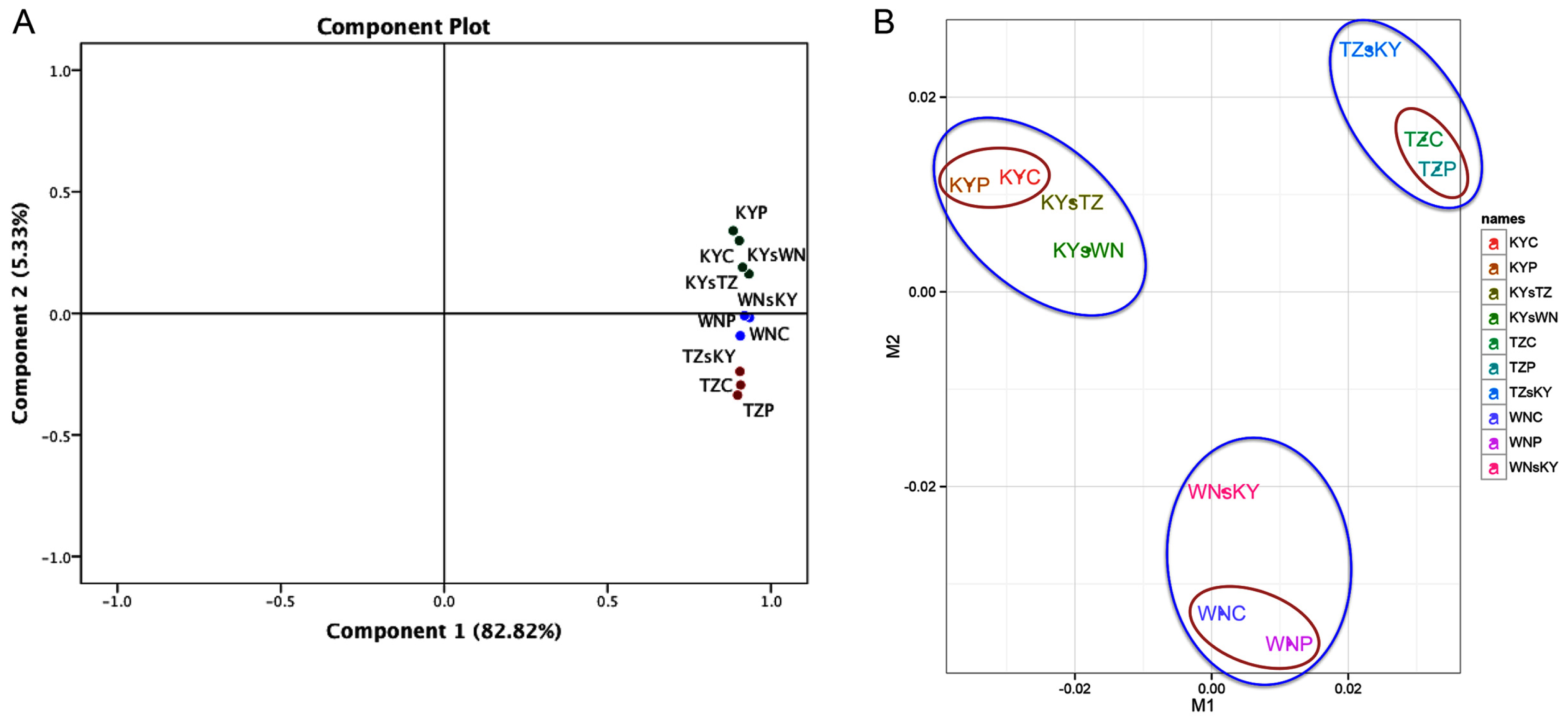
2.1. RNA-Seq Data Analyses
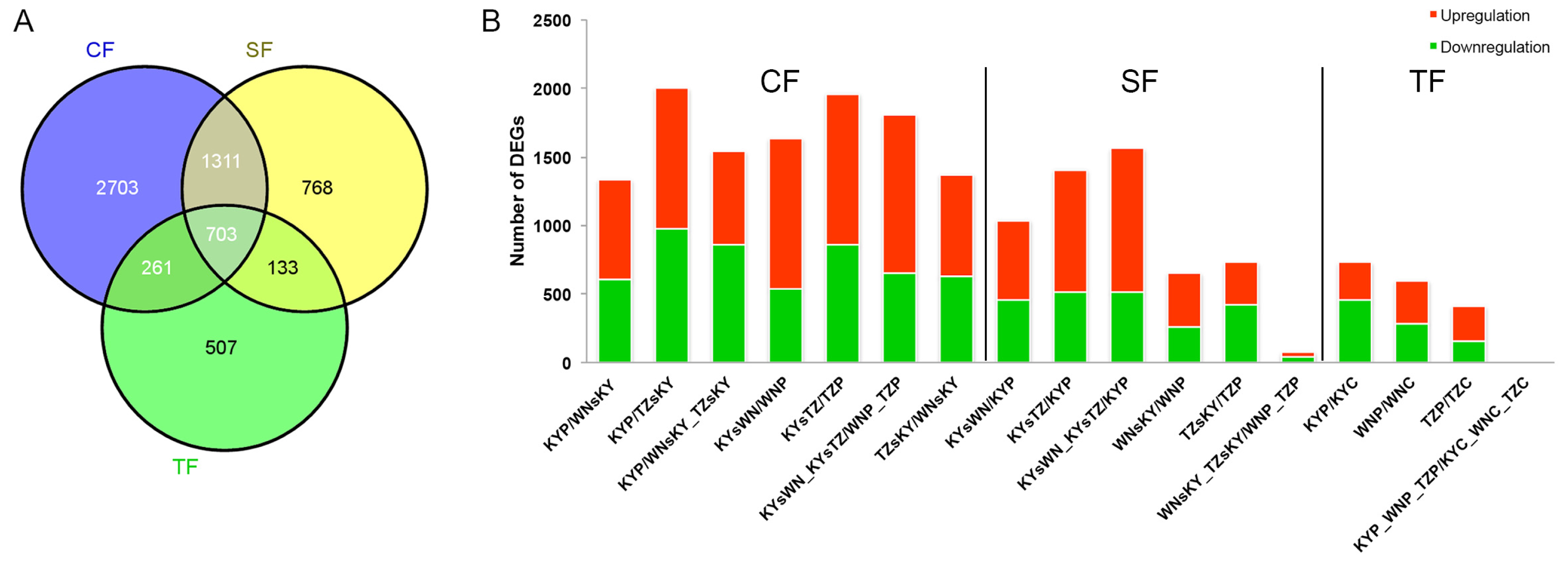
2.2. General Trend of DEGs in Tobacco Leaves
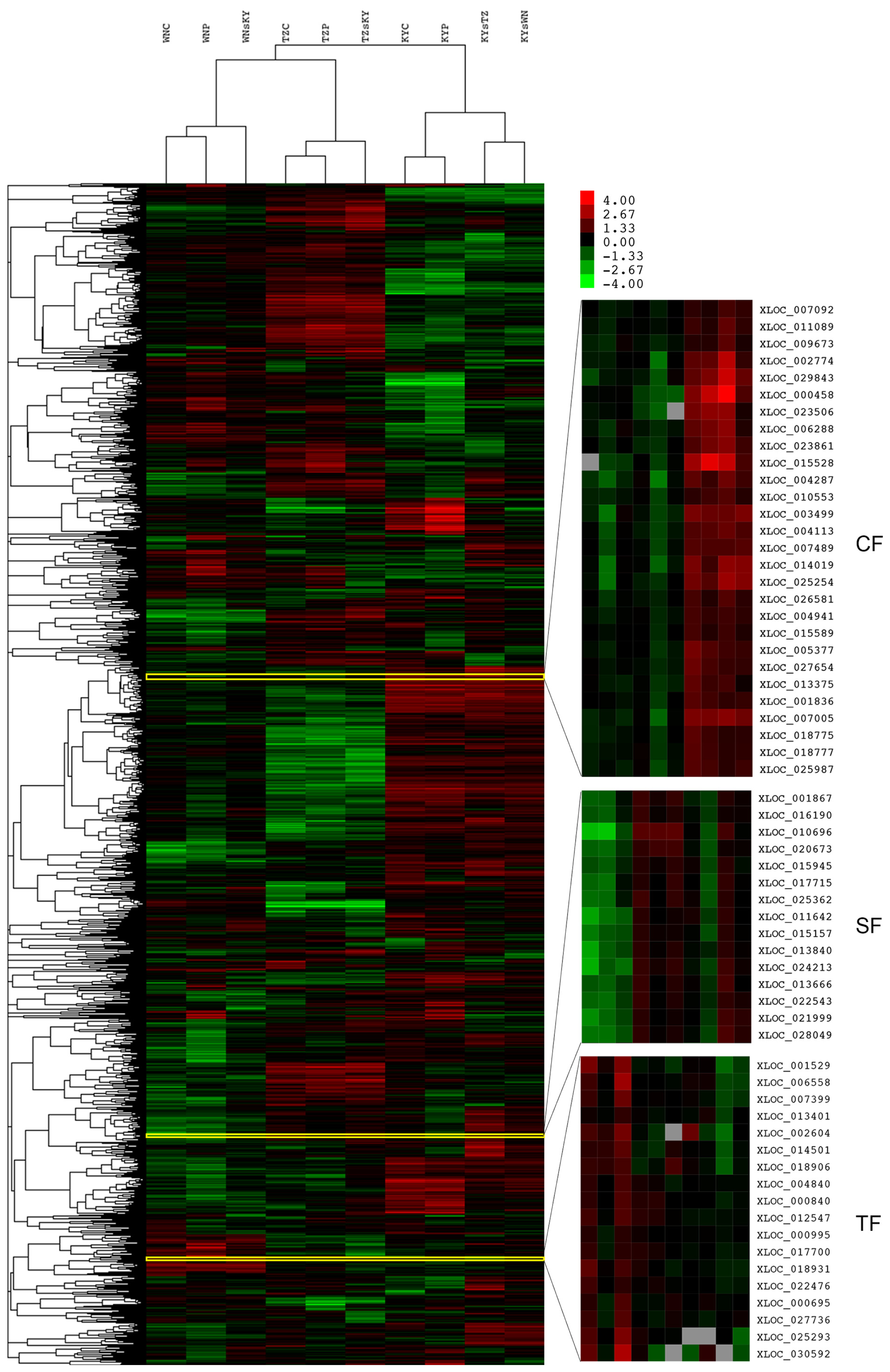
2.3. Functional Analysis of Common DEGs Affected by CFs, SFs, and TFs
2.4. Functional Analysis of CF-Specific DEGs
2.5. Functional Analysis of SF-Specific DEGs
2.6. Functional Analysis of TF-Specific DEGs
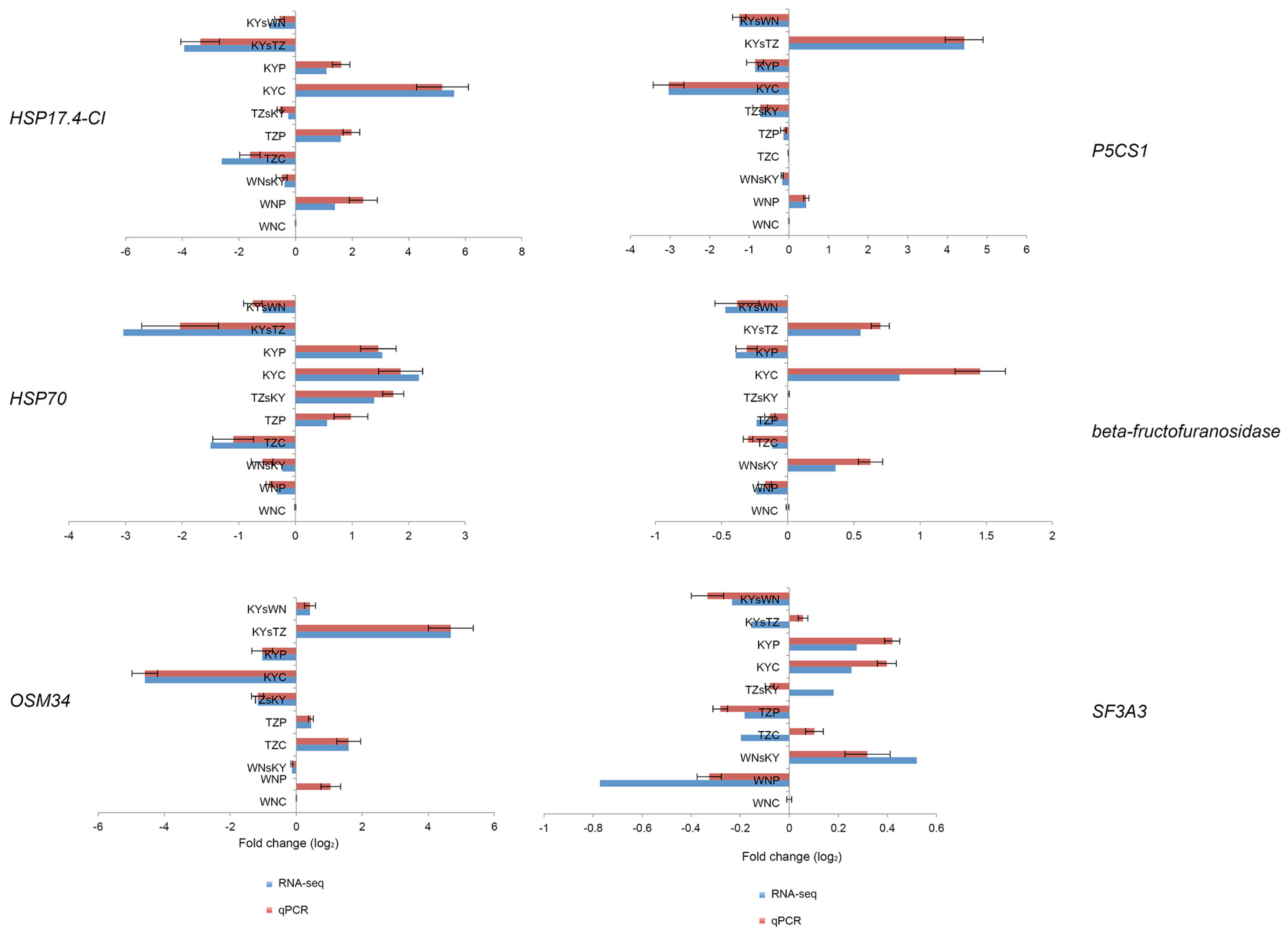
2.7. Validation of RNA-Seq Data by qRT-PCR
3. Experimental Section
3.1. Plant Materials
3.2. RNA Preparation
3.3. RNA-Seq Library Preparation and Sequencing
3.4. RNA-Seq Analysis
3.5. Quantitative Real-Time PCR (qRT-PCR) Validation
3.6. Measurement of Agronomic Traits and SOD, CAT and POD Activities of Tobacco Leaves
4. Conclusions
Acknowledgments
Conflicts of Interest
- Author ContributionsLB, LK, and DF carried out the experiments, analyzed the data, and drafted the manuscript. ZK contributed to data analysis. RZ, CY, ZH, and ZL performed RNA-seq, analyzed sequencing data. QC, GW and WJ performed experiments and reviewed the manuscript. PW designed the study and participated in writing the manuscript.
References
- Baker, N.T.; Capel, P.D. Environmental Factors that Influence the Location of Crop Agriculture in the Conterminous United States; U.S. Geological Survey: Reston, VA, USA, 2011; p. 72. [Google Scholar]
- Criddle, R.S.; Hopkin, M.S.; McArthur, E.D.; Hansen, L.D. Plant distribution and the temperature coefficient of metabolism. Plant Cell Environ 1994, 17, 233–243. [Google Scholar]
- Hinsinger, P.; Brauman, A.; Devau, N.; Gérard, F.; Jourdan, C.; Laclau, J.P.; le Cadre, E.; Jaillard, B.; Plassard, C. Acquisition of phosphorus and other poorly mobile nutrients by roots. Where do plant nutrition models fail? Plant Soil 2011, 348, 29–61. [Google Scholar]
- Chivenge, P.; Vanlauwe, B.; Six, J. Does the combined application of organic and mineral nutrient sources influence maize productivity? A meta-analysis. Plant Soil 2011, 342, 1–30. [Google Scholar]
- Waters, B.M.; Sankaran, R.P. Moving micronutrients from the soil to the seeds: Genes and physiological processes from a biofortification perspective. Plant Sci 2011, 180, 562–574. [Google Scholar]
- Wang, Z.H.; Li, S.X.; Malhi, S. Effects of fertilization and other agronomic measures on nutritional quality of crops. J. Sci. Food Agric 2008, 88, 7–23. [Google Scholar]
- Nyborg, M.; Solberg, E.D.; Izaurralde, R.C.; Malhi, S.S.; Molina-Ayala, M. Influence of long-term tillage, straw and N fertilizer on barley yield, plant-N uptake and soil-N balance. Soil Till. Res 1995, 36, 165–174. [Google Scholar]
- Malhi, S.S.; Grant, C.A.; Johnston, A.M.; Gill, K.S. Nitrogen fertilization management for no-till cereal production in the Canadian great plains: A review. Soil Till. Res 2001, 60, 101–122. [Google Scholar]
- Vakali, C.; Zaller, J.G.; Köpke, U. Reduced tillage effects on soil properties and growth of cereals and associated weeds under organic farming. Soil Till Res 2011, 111, 133–141. [Google Scholar]
- Rizhsky, L.; Hongjian, L.; Mittler, R. The combined effect of drought stress and heat shock on gene expression in tobacco. Plant Physiol 2002, 130, 1143–1151. [Google Scholar]
- Rizhsky, L.; Liang, H.; Shuman, J.; Shulaev, V.; Davletova, S.; Mittler, R. When defense pathways collide. The response of Arabidopsis to a combination of drought and heat stress. Plant Physiol 2004, 134, 1683–1696. [Google Scholar]
- Dal Santo, S.; Tornielli, G.B.; Zenoni, S.; Fasoli, M.; Farina, L.; Anesi, A.; Guzzo1, F.; Delledonne, M.; Pezzotti, M. The plasticity of the grapevine berry transcriptome. Genome Biol 2013, 14, R54. [Google Scholar]
- Tsirigos, A.; Haiminen, N.; Bilal, E.; Utro, F. GenomicTools: A computational platform for developing high-throughput analytics in genomics. Bioinformatics 2012, 28, 282–283. [Google Scholar]
- Kakumanu, A.; Ambavaram, M.M.R.; Klumas, C.; Krishnan, A.; Batlang, U. Effects of drought on gene expression in maize reproductive and leaf meristem tissue revealed by RNA-Seq. Plant Physiol 2012, 160, 846–867. [Google Scholar]
- O’Rourke, J.A.; Yang, S.S.; Miller, S.S.; Bucciarelli, B.; Liu, J.; Rydeen, A.; Bozsoki, Z.; Uhde-Stone, C.; Tu, Z.J.; Allan, D.; et al. An RNA-Seq transcriptome analysis of Pi deficient white lupin reveals novel insights into phosphorus acclimation in plants. Plant Physiol 2013, 161, 705–724. [Google Scholar]
- Dugas, D.; Monaco, M.; Olson, A.; Klein, R.; Kumari, S.; Ware, D.; Klein, P. Functional annotation of the transcriptome of sorghum bicolor in response to osmotic stress and abscisic acid. BMC Genomics 2011, 12, 514. [Google Scholar]
- Jaeger, P.A.; Doherty, C.; Ideker, T. Modeling transcriptome dynamics in a complex world. Cell 2012, 151, 1161–1162. [Google Scholar]
- Lei, B.; Zhao, X.H.; Zhang, K.; Zhang, J.; Ren, W.; Ren, Z.; Chen, Y.; Zhao, H.N.; Pan, W.J.; Chen, W.; et al. Comparative transcriptome analysis of tobacco (Nicotiana tabacum) leaves to identify aroma compound-related genes expressed in different cultivated regions. Mol. Biol. Rep 2013, 40, 345–357. [Google Scholar]
- Ali Reza, F. The effect of nitrogen and potassium fertilizer on yield, quality and some quantitative characteristics of flue-cured tobacco cv. Coker347. Afr. J. Agric. Res 2012, 7, 1827–1833. [Google Scholar]
- Orlando, F. Growth and development responses of tobacco (Nicotiana tabacum L.) to changes in physical and hydrological soil properties due to minimum tillage. Am. J. Plant Sci 2011, 2, 334–344. [Google Scholar]
- Bombarely, A.; Menda, N.; Tecle, I.Y.; Buels, R.M.; Strickler, S.; Fischer-York, T.; Pujar, A.; Leto, J.; Gosselin, J.; Mueller, L.A. The sol genomics network (solgenomics. net): Growing tomatoes using Perl. Nucleic Acids Res 2011, 39, D1149–D1155. [Google Scholar]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc 2012, 7, 562–578. [Google Scholar]
- Wang, Y.; Ghaffari, N.; Johnson, C.D.; Braga-Neto, U.M.; Wang, H.; Chen, R.; Zhou, H. Evaluation of the coverage and depth of transcriptome by RNA-Seq in chickens. BMC Bioinf 2011, 12, S5. [Google Scholar]
- Malone, J.H.; Oliver, B. Microarrays, deep sequencing and the true measure of the transcriptome. BMC Biol 2011, 9, 34. [Google Scholar]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol 2010, 28, 511–515. [Google Scholar]
- Edwards, K.D.; Bombarely, A.; Story, G.W.; Allen, F.; Mueller, L.A.; Coates, S.A.; Jones, L. TobEA: An atlas of tobacco gene expression from seed to senescence. BMC Genomics 2010, 11, 142. [Google Scholar]
- Chen, W.; Xiong, J.; Chen, Y.; Pan, W.J.; Li, Z.Y. Effects of climate and soil on the carotenoid and cuticular extract content of cured tobacco leaves. Acta Ecol. Sin 2013, 33, 3865–3877. [Google Scholar]
- Yoshida, T.; Fujita, Y.; Sayama, H.; Kidokoro, S.; Maruyama, K.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J 2010, 61, 672–685. [Google Scholar]
- Xie, D. COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 1998, 280, 1091–1094. [Google Scholar]
- Ann, T.; Kono, N.; Kosemura, S.; Yamahura, S.; Hasegawa, K. Isolation and characterization of an auxin-inducible SAUR gene from radish seedlings. Mitochondrial DNA 1998, 9, 329–333. [Google Scholar]
- Ha, S.; Vankova, R.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.S. Cytokinins: Metabolism and function in plant adaptation to environmental stresses. Trends Plant Sci 2012, 17, 172–179. [Google Scholar]
- Nagano, A.J.; Sato, Y.; Mihara, M.; Antonio, B.A.; Motoyama, R.; Itoh, H.; Nagamura, Y.; Izawa, T. Deciphering and prediction of transcriptome dynamics under fluctuating field conditions. Cell 2012, 151, 1358–1369. [Google Scholar]
- Sawa, M.; Kay, S.A. GIGANTEA directly activates Flowering Locus T inArabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2011, 108, 11698–11703. [Google Scholar]
- Mizoguchi, T.; Wheatley, K.; Hanzawa, Y.; Wright, L.; Mizoguchi, M.; Song, H.-R.; Carré, I.A.; Coupland, G. LHY and CCA1 are partially redundant genes required to maintain circadian rhythms inArabidopsis. Dev. Cell 2002, 2, 629–641. [Google Scholar]
- Dong, M.A.; Farré, E.M.; Thomashow, M.F. Circadian clock-associated 1 and late elongated hypocotyl regulate expression of the C-repeat binding factor (CBF) pathway inArabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 7241–7246. [Google Scholar]
- Nakamichi, N.; Kiba, T.; Henriques, R.; Mizuno, T.; Chua, N.H.; Sakakibara, H. Pseudo-response regulators 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 2010, 22, 594–605. [Google Scholar]
- Wang, Z.; Hu, H.; Huang, H.; Duan, K.; Wu, Z.; Wu, P. Regulation of OsSPX1 and OsSPX3 on expression of OsSPX domain genes and Pi-starvation signaling in rice. J. Integr. Plant Biol 2009, 51, 663–674. [Google Scholar]
- Chen, S.; Xia, G.; Zhao, W.; Wu, F.; Zhang, G. Characterization of leaf photosynthetic properties for no-tillage rice. Rice Sci 2007, 14, 283–288. [Google Scholar]
- Lu, K.; Chai, Y.R.; Zhang, K.; Wang, R.; Chen, L.; Lei, B.; Lu, J.; Xu, X.F.; Li, J.N. Cloning and characterization of phosphorus starvation inducible Brassica napus PURPLE ACID PHOSPHATASE 12 gene family, and imprinting of a recently evolved MITE-minisatellite twin structure. Theor. Appl. Genet 2008, 117, 963–975. [Google Scholar]
- De Hoon, M.J.; Imoto, S.; Nolan, J.; Miyano, S. Open source clustering software. Bioinformatics 2004, 20, 1453–1454. [Google Scholar]
- Saldanha, A.J. Java Treeview—Extensible visualization of microarray data. Bioinformatics 2004, 20, 3246–3248. [Google Scholar]
- Lamesch, P.; Berardini, T.Z.; Li, D.; Swarbreck, D.; Wilks, C.; Sasidharan, R.; Muller, R.; Dreher, K.; Alexander, D.L.; Garcia-Hernandez, M. The Arabidopsis information resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res 2012, 40, D1202–D1210. [Google Scholar]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res 1997, 25, 3389–3402. [Google Scholar]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar]
- Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.L.; Ideker, T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics 2011, 27, 431–432. [Google Scholar]
- Maere, S.; Heymans, K.; Kuiper, M. Bingo: A cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 2005, 21, 3448–3449. [Google Scholar]
- KOBAS 2.0. Available online: http://kobas.cbi.pku.edu.cn/home.do (accessed on 4 March 2014).
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res 2011, 39, W316–W322. [Google Scholar]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 1995, 289–300. [Google Scholar]
- Goff, L.; Trapnell, C.; Kelley, D. CummeRbund: Analysis, Exploration, Manipulation, and Visualization of Cufflinks High-Throughput Sequencing Data. 8 May 2013.
- Rozen, S.; Skaletsky, H. Primer3 on the www for general users and for biologist programmers. In Bioinformatics Bethods and Protocols: Methods in Molecular Biology; Krawetz, S., Misener, S., Eds.; Humana Press: Totowa, NJ, USA, 2000; Volume 132, pp. 365–386. [Google Scholar]
- Schmidt, G.W.; Delaney, S.K. Stable internal reference genes for normalization of real-time RT-PCR in tobacco (Nicotiana tabacum) during development and abiotic stress. Mol. Genet. Genomics 2010, 283, 233–241. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar]
- Ranieri, A.; Petacco, F.; Castagna, A.; Soldatini, G.F. Redox state and peroxidase system in sunflower plants exposed to ozone. Plant Sci 2000, 159, 159–167. [Google Scholar]
| Sample | Total reads | Mapped reads | % Mapped reads | Number of transcripts |
|---|---|---|---|---|
| KYC | 6,010,205 | 4,164,387 | 69.29% | 26,864 |
| KYP | 6,127,551 | 4,276,322 | 69.79% | 25,823 |
| KYsWN | 5,566,935 | 3,894,388 | 69.96% | 25,001 |
| KYsTZ | 6,020,409 | 4,162,982 | 69.15% | 25,550 |
| WNC | 5,252,135 | 3,663,806 | 69.76% | 23,971 |
| WNP | 5,913,266 | 4,259,827 | 72.04% | 24,185 |
| WNsKY | 5,218,808 | 3,704,089 | 70.98% | 24,261 |
| TZC | 6,091,837 | 4,254,123 | 69.83% | 25,128 |
| TZP | 6,005,103 | 4,222,155 | 70.31% | 25,540 |
| TZsKY | 6,260,204 | 4,455,821 | 71.18% | 26,100 |
| Overall | 58,466,453 | 41,057,900 | 70.22% | 31,057 |
| Group | No. | Combination | Number of DR genes | Number of UR genes | Number of DEGs |
|---|---|---|---|---|---|
| (i) Different CFs and the same SFs and TFs | 1 | KYP/WNsKY | 610 | 722 | 1332 |
| 2 | KYP/TZsKY | 971 | 1029 | 2000 | |
| 3 | KYP/WNsKY_TZsKY | 858 | 679 | 1537 | |
| 4 | KYsWN/WNP | 533 | 1100 | 1633 | |
| 5 | KYsTZ/TZP | 857 | 1104 | 1961 | |
| 6 | KYsWN_KYsTZ/WNP_TZP | 648 | 1155 | 1803 | |
| 7 | TZsKY/WNsKY | 631 | 739 | 1370 | |
| (ii) The same CFs and different SFs and TFs | 8 | KYsWN/KYC | 505 | 330 | 835 |
| 9 | KYsTZ/KYC | 654 | 585 | 1239 | |
| 10 | KYsWN_KYsTZ/KYC | 558 | 604 | 1162 | |
| 11 | WNsKY/WNC | 164 | 242 | 406 | |
| 12 | TZsKY/TZC | 331 | 513 | 844 | |
| 13 | WNsKY_TZsKY/WNC_TZC | 8 | 18 | 26 | |
| (iii) Different SFs and the same CFs and TFs | 14 | KYsWN/KYP | 462 | 576 | 1038 |
| 15 | KYsTZ/KYP | 514 | 893 | 1407 | |
| 16 | KYsWN_KYsTZ/KYP | 518 | 1041 | 1559 | |
| 17 | WNsKY/WNP | 255 | 395 | 650 | |
| 18 | TZsKY/TZP | 416 | 315 | 731 | |
| 19 | WNsKY_TZsKY/WNP_TZP | 44 | 30 | 74 | |
| (iv) The same SFs and different CFs and TFs | 20 | WNsKY/KYC | 960 | 506 | 1466 |
| 21 | TZsKY/KYC | 1168 | 737 | 1905 | |
| 22 | WNsKY_TZsKY/KYC | 662 | 543 | 1205 | |
| 23 | KYsWN/WNC | 362 | 850 | 1212 | |
| 24 | KYsTZ/TZC | 668 | 1143 | 1811 | |
| 25 | KYsWN_KYsTZ/WNC_TZC | 317 | 709 | 1026 | |
| (v) Different TFs and the same CFs and SFs; | 26 | KYP/KYC | 452 | 277 | 729 |
| 27 | WNP/WNC | 289 | 307 | 596 | |
| 28 | TZP/TZC | 151 | 259 | 410 | |
| 29 | KYP_WNP_TZP/KYC_WNC_TZC | 0 | 0 | 0 | |
| (vi) The same TFs and different CFs and SFs | 30 | KYC/WNC | 523 | 1212 | 1735 |
| 31 | KYC/TZC | 710 | 1412 | 2122 | |
| 32 | KYC/WNC_TZC | 531 | 794 | 1325 | |
| 33 | TZC/WNC | 625 | 821 | 1446 | |
| 34 | KYP/WNP | 750 | 1082 | 1832 | |
| 35 | KYP/TZP | 1034 | 1109 | 2143 | |
| 36 | KYP/WNP_TZP | 1144 | 1065 | 2209 | |
| 37 | TZP/WNP | 529 | 956 | 1485 | |
| Location | Treatments | 45 DAT | 70 DAT | ||||
|---|---|---|---|---|---|---|---|
| CAT | POD | SOD | CAT | POD | SOD | ||
| KY | KYC | 140.8 | 1551.3 | 611.7 | 101.4 | 6298.3 | 481.3 |
| KYP | 128.0 | 2181.9 | 648.2 | 93.5 | 6914.7 | 483.9 | |
| KYsTZ | 116.4 | 4275.2 | 594.0 | 108.8 | 8468.2 | 545.4 | |
| KYsWN | 124.5 | 3929.5 | 654.1 | 74.7 | 9298.9 | 454.1 | |
| TZ | TZC | 378.7 | 3929.3 | 511.5 | 94.3 | 4362.9 | 500.7 |
| TZP | 391.9 | 4697.0 | 476.8 | 134.3 | 4474.2 | 449.5 | |
| TZsKY | 361.2 | 3435.4 | 514.5 | 175.9 | 3724.5 | 499.4 | |
| WN | WNC | 175.7 | 1780.9 | 509.4 | 142.4 | 5180.0 | 516.9 |
| WNP | 177.7 | 2255.6 | 469.8 | 144.2 | 6428.5 | 470.8 | |
| WNsKY | 208.3 | 1426.6 | 507.7 | 143.5 | 4348.7 | 492.8 | |
| GO-ID | p-value | Input number | Background number | Description |
|---|---|---|---|---|
| 0048578 | 4.00 × 109 | 8 | 8 | positive regulation of long-day photoperiodism, flowering |
| 0010378 | 4.00 × 109 | 8 | 8 | temperature compensation of the circadian clock |
| 0009813 | 7.20 × 109 | 20 | 52 | flavonoid biosynthetic process |
| 0042398 | 1.51 × 108 | 44 | 200 | cellular amino acid derivative biosynthetic process |
| 0055114 | 5.23 × 108 | 237 | 1916 | oxidation reduction |
| 0010229 | 1.08 × 107 | 11 | 19 | inflorescence development |
| 0006575 | 2.18 × 107 | 57 | 316 | cellular amino acid derivative metabolic process |
| 0009812 | 2.20 × 107 | 20 | 62 | flavonoid metabolic process |
| 0009699 | 4.81 × 107 | 23 | 82 | phenylpropanoid biosynthetic process |
| 0048586 | 5.16 × 107 | 8 | 11 | regulation of long-day photoperiodism, flowering |
| 0006857 | 7.65 × 107 | 17 | 50 | oligopeptide transport |
| 0015833 | 7.65 × 107 | 17 | 50 | peptide transport |
| 0009698 | 9.35 × 107 | 29 | 123 | phenylpropanoid metabolic process |
| 0019748 | 1.01 × 106 | 44 | 230 | secondary metabolic process |
| 0006355 | 1.57 × 106 | 161 | 1262 | regulation of transcription, DNA-dependent |
| 0045449 | 1.99 × 106 | 161 | 1267 | regulation of transcription |
| 0051252 | 2.52 × 106 | 161 | 1272 | regulation of RNA metabolic process |
| 0008215 | 3.27 × 106 | 6 | 7 | spermine metabolic process |
| 0006597 | 3.27 × 106 | 6 | 7 | spermine biosynthetic process |
| 0010556 | 6.96 × 106 | 162 | 1304 | regulation of macromolecule biosynthetic process |
| 0008295 | 7.34 × 106 | 8 | 14 | spermidine biosynthetic process |
| 0031326 | 8.48 × 106 | 166 | 1347 | regulation of cellular biosynthetic process |
| 0009889 | 1.05 × 105 | 166 | 1352 | regulation of biosynthetic process |
| 0008216 | 1.45 × 105 | 8 | 15 | spermidine metabolic process |
| 0006835 | 2.38 × 105 | 7 | 12 | dicarboxylic acid transport |
| 0051171 | 2.62 × 105 | 167 | 1384 | regulation of nitrogen compound metabolic process |
| 0019219 | 2.89 × 105 | 165 | 1367 | regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process |
| 0015798 | 3.15 × 105 | 5 | 6 | myo-inositol transport |
| 0006596 | 6.03 × 105 | 9 | 22 | polyamine biosynthetic process |
| 0006833 | 6.17 × 105 | 11 | 32 | water transport |
| 0042044 | 6.17 × 105 | 11 | 32 | fluid transport |
| 0006725 | 9.44 × 105 | 52 | 341 | cellular aromatic compound metabolic process |
| 0051258 | 1.06 × 104 | 13 | 45 | protein polymerization |
| 0006595 | 1.79 × 104 | 10 | 30 | polyamine metabolic process |
| 0080090 | 1.82 × 104 | 170 | 1466 | regulation of primary metabolic process |
| 0000160 | 1.91 × 104 | 25 | 130 | two-component signal transduction system (phosphorelay) |
| 0015791 | 2.52 × 104 | 5 | 8 | polyol transport |
| 0015850 | 2.52 × 104 | 5 | 8 | organic alcohol transport |
| 0009610 | 2.95 × 104 | 4 | 5 | response to symbiotic fungus |
| 0009873 | 3.36 × 104 | 13 | 50 | ethylene mediated signaling pathway |
| 0010468 | 3.41 × 104 | 162 | 1405 | regulation of gene expression |
| 0019438 | 3.46 × 104 | 33 | 198 | aromatic compound biosynthetic process |
| 0006629 | 3.87 × 104 | 98 | 784 | lipid metabolic process |
| 0042752 | 5.76 × 104 | 8 | 23 | regulation of circadian rhythm |
| 0008610 | 6.35 × 104 | 58 | 422 | lipid biosynthetic process |
| 0008202 | 6.75 × 104 | 12 | 47 | steroid metabolic process |
| 0051552 | 7.97 × 104 | 8 | 24 | flavone metabolic process |
| 0051553 | 7.97 × 104 | 8 | 24 | flavone biosynthetic process |
| 0051554 | 7.97 × 104 | 8 | 24 | flavonol metabolic process |
| 0051555 | 7.97 × 104 | 8 | 24 | flavonol biosynthetic process |
| 0035235 | 8.07 × 104 | 6 | 14 | ionotropic glutamate receptor signaling pathway |
| 0007215 | 8.07 × 104 | 6 | 14 | glutamate signaling pathway |
| 0060255 | 8.65 × 104 | 163 | 1444 | regulation of macromolecule metabolic process |
| 0071369 | 9.03 × 104 | 13 | 55 | cellular response to ethylene stimulus |
| 0009409 | 9.30 × 104 | 42 | 286 | response to cold |
| 0031323 | 9.94 × 104 | 184 | 1661 | regulation of cellular metabolic process |
| 0009755 | 1.12 × 103 | 45 | 315 | hormone-mediated signaling pathway |
| 0032870 | 1.13 × 103 | 46 | 324 | cellular response to hormone stimulus |
| 0042401 | 1.22 × 103 | 12 | 50 | cellular biogenic amine biosynthetic process |
| 0071495 | 1.22 × 103 | 49 | 352 | cellular response to endogenous stimulus |
| 0046148 | 1.36 × 103 | 21 | 116 | pigment biosynthetic process |
| 0010033 | 1.52 × 103 | 116 | 993 | response to organic substance |
| 0009723 | 1.55 × 103 | 22 | 125 | response to ethylene stimulus |
| 0009608 | 1.65 × 103 | 5 | 11 | response to symbiont |
| GO-ID | p-value | Input number | Background number | Description |
|---|---|---|---|---|
| 0016036 | 3.89 × 105 | 8 | 52 | cellular response to phosphate starvation |
| 0000041 | 8.71 × 105 | 9 | 74 | transition metal ion transport |
| 0007154 | 1.10 × 104 | 15 | 195 | cell communication |
| 0009765 | 1.97 × 104 | 8 | 65 | photosynthesis, light harvesting |
| 0009867 | 2.73 × 104 | 6 | 37 | jasmonic acid mediated signaling pathway |
| 0071395 | 2.73 × 104 | 6 | 37 | cellular response to jasmonic acid stimulus |
| 0006464 | 3.34 × 104 | 75 | 2049 | protein modification process |
| 0035303 | 4.11 × 104 | 4 | 15 | regulation of dephosphorylation |
| 0006950 | 4.38 × 104 | 79 | 2205 | response to stress |
| 0050896 | 5.30 × 104 | 119 | 3644 | response to stimulus |
| 0006664 | 5.55 × 104 | 6 | 42 | glycolipid metabolic process |
| 0009875 | 5.57 × 104 | 7 | 58 | pollen-pistil interaction |
| 0006827 | 6.15 × 104 | 2 | 2 | high-affinity iron ion transport |
| 0046506 | 6.15 × 104 | 2 | 2 | sulfolipid biosynthetic process |
| 0046505 | 6.15 × 104 | 2 | 2 | sulfolipid metabolic process |
| 0006825 | 6.68 × 104 | 5 | 29 | copper ion transport |
| 0009247 | 7.85 × 104 | 5 | 30 | glycolipid biosynthetic process |
| 0009267 | 8.20 × 104 | 8 | 80 | cellular response to starvation |
| 0009743 | 8.66 × 104 | 12 | 165 | response to carbohydrate stimulus |
| 0043687 | 9.48 × 104 | 68 | 1883 | post-translational protein modification |
| GO-ID | p-value | Input number | Background number | Description |
|---|---|---|---|---|
| 0015833 | 1.28 × 104 | 6 | 50 | peptide transport |
| 0042454 | 1.29 × 104 | 3 | 7 | ribonucleoside catabolic process |
| Cultivated regions | CF | SF | TF |
|---|---|---|---|
| KY | KYC | KYsTZ and KYsWN | KYP |
| TZ | TZC | TZsKY | TZP |
| WN | WNC | WNsKY | WNP |
© 2014 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Lei, B.; Lu, K.; Ding, F.; Zhang, K.; Chen, Y.; Zhao, H.; Zhang, L.; Ren, Z.; Qu, C.; Guo, W.; et al. RNA Sequencing Analysis Reveals Transcriptomic Variations in Tobacco (Nicotiana tabacum) Leaves Affected by Climate, Soil, and Tillage Factors. Int. J. Mol. Sci. 2014, 15, 6137-6160. https://doi.org/10.3390/ijms15046137
Lei B, Lu K, Ding F, Zhang K, Chen Y, Zhao H, Zhang L, Ren Z, Qu C, Guo W, et al. RNA Sequencing Analysis Reveals Transcriptomic Variations in Tobacco (Nicotiana tabacum) Leaves Affected by Climate, Soil, and Tillage Factors. International Journal of Molecular Sciences. 2014; 15(4):6137-6160. https://doi.org/10.3390/ijms15046137
Chicago/Turabian StyleLei, Bo, Kun Lu, Fuzhang Ding, Kai Zhang, Yi Chen, Huina Zhao, Lin Zhang, Zhu Ren, Cunmin Qu, Wenjing Guo, and et al. 2014. "RNA Sequencing Analysis Reveals Transcriptomic Variations in Tobacco (Nicotiana tabacum) Leaves Affected by Climate, Soil, and Tillage Factors" International Journal of Molecular Sciences 15, no. 4: 6137-6160. https://doi.org/10.3390/ijms15046137
APA StyleLei, B., Lu, K., Ding, F., Zhang, K., Chen, Y., Zhao, H., Zhang, L., Ren, Z., Qu, C., Guo, W., Wang, J., & Pan, W. (2014). RNA Sequencing Analysis Reveals Transcriptomic Variations in Tobacco (Nicotiana tabacum) Leaves Affected by Climate, Soil, and Tillage Factors. International Journal of Molecular Sciences, 15(4), 6137-6160. https://doi.org/10.3390/ijms15046137