Construction and Analysis of Siberian Tiger Bacterial Artificial Chromosome Library with Approximately 6.5-Fold Genome Equivalent Coverage
Abstract
:1. Introduction
2. Results
2.1. Cell Cultures and Characteristic Tests
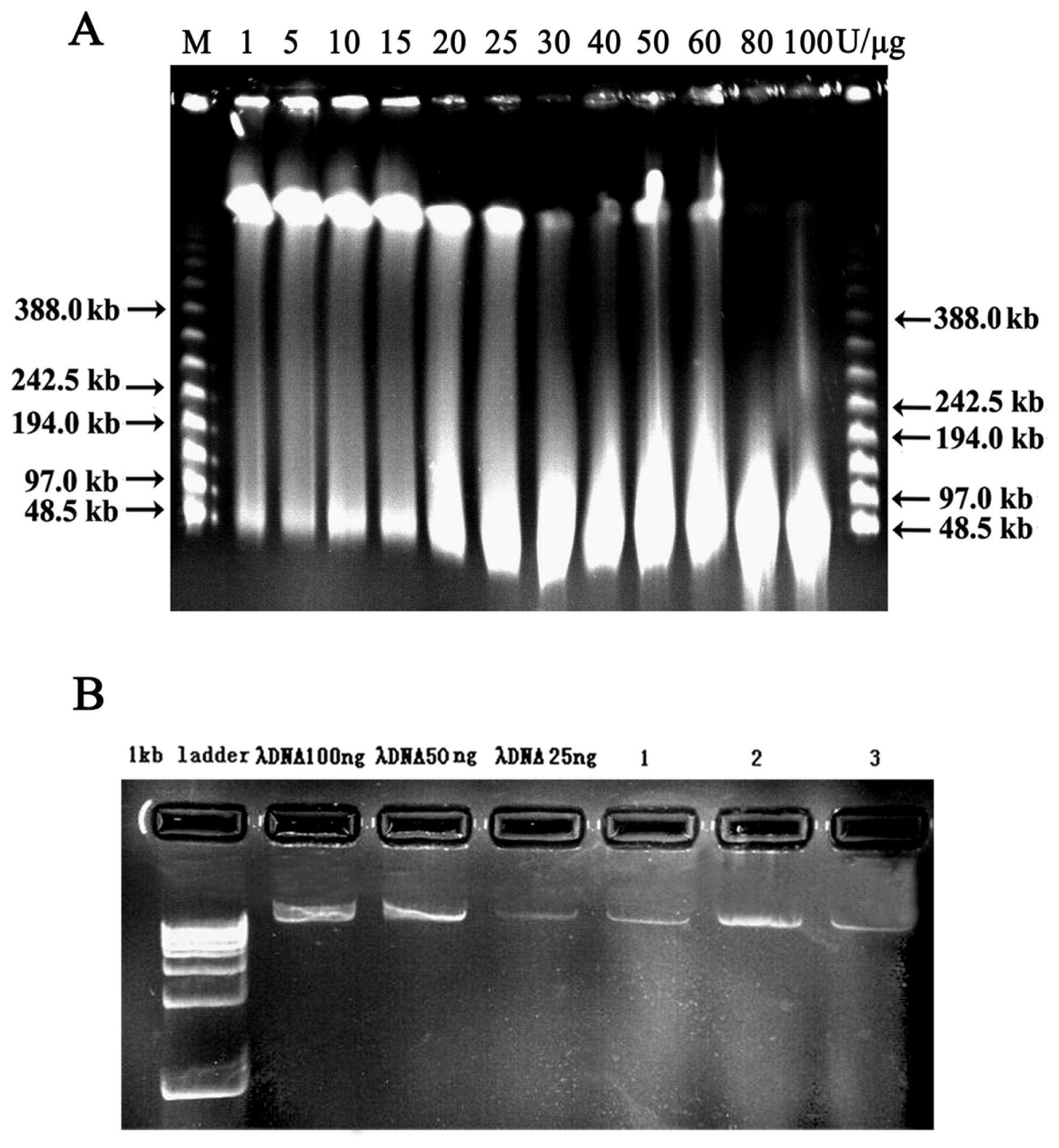
2.2. Vector Preparation and High-Molecular-Weight (HMW) Genomic DNA Preparation
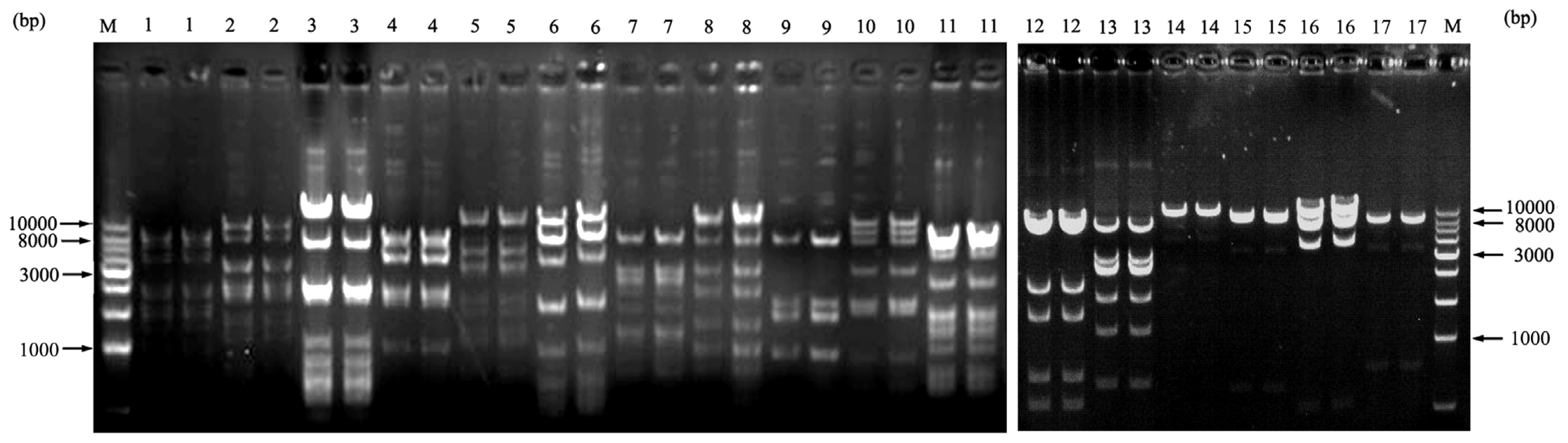
2.3. Characterization and Insert Size Testing of BAC Library
2.4. The Testing of Library Stability
2.5. BAC End Sequence Analysis
2.6. BAC Library Screening
3. Discussion
4. Materials and Methods
4.1. Preparation of BAC Vector
4.2. High-Molecular-Weight (HMW) Genomic DNA Preparation
4.3. Ligation and Transformation
4.4. Large-Scale BAC Clone Production
4.5. Insert Size Distribution of Siberian Tiger BAC Library
4.6. Library Pooling and BAC Library Screening
4.7. BAC End Sequencing
5. Conclusions
Acknowledgments
Conflicts of Interest
References
- Luo, M.; Kim, H.; Kudrna, D.; Sisneros, N.B.; Lee, S.J.; Mueller, C.; Collura, K.; Zuccolo, A.; Buckingham, E.B.; Grim, S.M.; et al. Construction of a nurse shark (Ginglymostoma cirratum) bacterial artificial chromosome (BAC) library and a preliminary genome survey. BMC Genomics 2006, 7, 106. [Google Scholar]
- Anistoroaei, R.; Ten Hallers, B.; Nefedov, M.; Christensen, K.; de Jong, P. Construction of an American mink bacterial artificial chromosome (BAC) library and sequencing candidate genes important for the fur industry. BMC Genomics 2011, 12, 354. [Google Scholar]
- Chen, R.; Sodergren, E.; Weinstock, G.M.; Gibbs, R.A. Dynamic building of a BAC clone tiling path for the rat genome sequencing project. Genome Res. 2004, 14, 679–684. [Google Scholar]
- Suetsugu, Y.; Minami, H.; Shimomura, M.; Sasanuma, S.; Narukawa, J.; Mita, K.; Yamamoto, K. End-sequencing and characterization of silkworm (Bombyx mori) bacterial artificial chromosome libraries. BMC Genomics 2007, 8, 314. [Google Scholar]
- Fujisaki, S.; Mizoguchi, Y.; Takahashi, S.; Chen, Y.Z.; Suzuki, K.; Asakawa, S.; Soeda, E.; Shimizu, N.; Sugimoto, Y.; Yasue, H. Construction of a bovine bacterial artificial chromosome library from fibroblasts used for cloned cattle. Anim. Genet. 2002, 33, 379–381. [Google Scholar]
- Stafuzza, N.B.; Abbey, C.A.; Gill, C.A.; Womack, J.E.; Amaral, M.E. Construction and preliminary characterization of a river buffalo bacterial artificial chromosome library. Genet. Mol. Res. 2012, 11, 3013–3019. [Google Scholar]
- Schibler, L.; Vaiman, D.; Oustry, A.; Guinec, N.; Dangy-Caye, A.L.; Billault, A.; Cribiu, E.P. Construction and extensive characterization of a goat bacterial artificial chromosome library with threefold genome coverage. Mamm. Genome 1998, 9, 119–124. [Google Scholar]
- Godard, S.; Schibler, L.; Oustry, A.; Cribiu, E.P.; Guerin, G. Construction of a horse BAC library and cytogenetical assignment of 20 type I and type II markers. Mamm. Genome 1998, 9, 633–637. [Google Scholar]
- Jeon, J.T.; Park, E.W.; Jeon, H.J.; Kim, T.H.; Lee, K.T.; Cheong, I.C. A large-insert porcine library with sevenfold genome coverage: A tool for positional cloning of candidate genes for major quantitative traits. Mol. Cells 2003, 16, 113–116. [Google Scholar]
- Liu, C.; Guo, Y.; Lu, T.; Wu, H.; Na, R.; Li, X.; Guan, W.; Ma, Y. Construction and preliminary characterization analysis of Wuzhishan miniature pig bacterial artificial chromosome library with approximately 8-fold genome equivalent coverage. BioMed. Res. Int. 2013, 2013, 587493. [Google Scholar]
- Vaiman, D.; Billault, A.; Tabet-Aoul, K.; Schibler, L.; Vilette, D.; Oustry-Vaiman, A.; Soravito, C.; Cribiu, E.P. Construction and characterization of a sheep BAC library of three genome equivalents. Mamm. Genome 1999, 10, 585–587. [Google Scholar]
- Liu, W.; Zhao, Y.; Liu, Z.; Zhang, Y.; Lian, Z.; Li, N. Construction of a 7-fold BAC library and cytogenetic mapping of 10 genes in the giant panda (Ailuropoda melanoleuca). BMC Genomics 2006, 7, 294. [Google Scholar]
- Luo, S.J.; Kim, J.H.; Johnson, W.E.; van der Walt, J.; Martenson, J.; Yuhki, N.; Miquelle, D.G.; Uphyrkina, O.; Goodrich, J.M.; Quigley, H.B.; et al. Phylogeography and genetic ancestry of tigers (Panthera tigris). PLoS Biol. 2004, 2, e442. [Google Scholar]
- Liu, C.Q.; Lu, T.F.; Feng, B.G.; Liu, D.; Guan, W.J.; Ma, Y.H. Construction of cDNA library and preliminary analysis of expressed sequence tags from Siberian tiger. Int. J. Biol. Sci. 2010, 6, 584–589. [Google Scholar]
- Liu, C.; Liu, D.; Guo, Y.; Lu, T.; Li, X.; Zhang, M.; Ma, J.; Ma, Y.; Guan, W. Construction of a full-length enriched cDNA library and preliminary analysis of expressed sequence tags from Bengal tiger Panthera tigris. Int. J. Mol. Sci. 2013, 14, 11072–11083. [Google Scholar]
- Hu, Y.; Lu, Y.; Ma, D.; Guo, W.; Zhang, T. Construction and characterization of a bacterial artificial chromosome library for the A-genome of cotton (G arboreum L). J. Biomed. Biotechnol. 2010, 2010, 457137:1–457137:5. [Google Scholar]
- Liu, C.Q.; Bai, C.Y.; Guo, Y.; Liu, D.; Guan, W.J.; Ma, Y.H. End-sequencing and characterization of Siberian Tiger (Panthera tigris altaica) bacterial artificial chromosome libraries. Int. J. Mol. Sci. 2014. to be submitted for publication. [Google Scholar]
- Han, Y.; Korban, S.S. An overview of the apple genome through BAC end sequence analysis. Plant Mol. Biol. 2008, 67, 581–588. [Google Scholar]
- Ling, F.; Wei, L.; Wang, T.; Chen, Y.; Zhu, X.; Li, J.; Liu, T.; Du, H.; Wang, H.; Wang, J. Cloning and characterization of the 5′-flanking region of the pig cocaine- and amphetamine-regulated transcript gene. DNA Cell Biol. 2011, 30, 91–97. [Google Scholar]
- Klukowska, J.; Szczerbal, I.; Wengi-Piasecka, A.; Switonski, M.; Schelling, C.; Gmur, A.; Dolf, G. Characterization and mapping of canine microsatellites isolated from BAC clones harbouring DNA sequences homologous to seven human genes. Anim. Genet. 2004, 35, 404–407. [Google Scholar]
- Airmet, K.W.; Hinckley, J.D.; Tree, L.T.; Moss, M.; Blumell, S.; Ulicny, K.; Gustafson, A.K.; Weed, M.; Theodosis, R.; Lehnardt, M.; et al. Construction of a llama bacterial artificial chromosome library with approximately 9-fold genome equivalent coverage. J. Biomed. Biotechnol. 2012, 2012, 371414. [Google Scholar]
- Qian, Y.; Jin, L.; Su, B. Construction and characterization of bacterial artificial chromosome library of black-handed spider monkey (Ateles geoffroyi). Genome 2004, 47, 239–245. [Google Scholar]
- Talbot, N.C.; Caperna, T.J.; Garrett, W.M. Growth and development symposium: Development characterization and use of a porcine epiblast-derived liver stem cell line: ARS-PICM-19. J. Anim. Sci. 2013, 91, 66–77. [Google Scholar]
- Yasui, G.S.; Fujimoto, T.; Sakao, S.; Yamaha, E.; Arai, K. Production of loach (Misgurnus anguillicaudatus) germ-line chimera using transplantation of primordial germ cells isolated from cryopreserved blastomeres. J. Anim. Sci. 2011, 89, 2380–2388. [Google Scholar]
- Osoegawa, K.; Woon, P.Y.; Zhao, B.; Frengen, E.; Tateno, M.; Catanese, J.J.; de Jong, P.J. An improved approach for construction of bacterial artificial chromosome libraries. Genomics 1998, 52, 1–8. [Google Scholar]


| Sub-library | Sample | Vector | Bacterial strain | Cloning enzyme | Total clones | Plate No. | Non-insert clone (%) | Average insert size a (kb) |
|---|---|---|---|---|---|---|---|---|
| BAC11-B | Blood cells | pBeloBAC11 | E. coli DH10B | HindIII | 92,160 | 1–240 | 2.1 (2) | 126.5 |
| pCC1-B | Blood cells | Copy Control™ pCC1 BAC™ | EPI300™-T1R E. coli | HindIII | 23,040 | 241–300 | 5.2 (5) | 116.7 |
| BAC11-F | Fibroblasts | pBeloBAC11 | E. coli DH10B | HindIII | 23,040 | 301–360 | 3.1 (3) | 114.3 |
| pCC1-F | Fibroblasts | Copy Control™ pCC1 BAC™ | EPI300™-T1R E. coli | HindIII | 15,360 | 361–400 | 1.0 (1) | 110.1 |
| Total | 153,600 | 400 | 2.6 | 116.5 |
| Marker name | Location Chr | Number of positive superpools | Number of positive clones |
|---|---|---|---|
| KF463190 | A1 | 4/2 | 6/2 |
| KF463196 | A2 | 3/1 | 4/1 |
| KF463200 | A3 | 3/0 | 9/3 |
| KF463203 | B1 | 2/0 | 5/2 |
| KF463210 | B2 | 1/2 | 4/2 |
| FCA201 | B3 | 3/2 | 4/3 |
| FCA069 | B4 | 3/1 | 4/1 |
| KF463223 | C1 | 3/NF | 6/0 |
| FCA043 | C2 | 3/1 | 5/2 |
| KF463232 | D1 | 4/1 | 6/1 |
| KF463233 | D2 | 2/2 | 4/4 |
| KF463238 | D3 | 2/3 | 5/3 |
| KF463238 | D4 | NF/2 | 0/2 |
| KF463245 | E1 | 4/1 | 8/1 |
| KF463247 | E2 | 3/1 | 5/2 |
| KF463252 | E3 | 3/2 | 5/2 |
| KF463255 | F1 | 2/NF | 4/1 |
| FCA094 | F2 | 4/2 | 5/1 |
| KF463257 | X | 2/1 | 3/1 |
| SRY | Y | 1/0 | 1/0 |
| Average number of positive clone | 4.90/1.84 |
© 2014 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Liu, C.; Bai, C.; Guo, Y.; Liu, D.; Lu, T.; Li, X.; Ma, J.; Ma, Y.; Guan, W. Construction and Analysis of Siberian Tiger Bacterial Artificial Chromosome Library with Approximately 6.5-Fold Genome Equivalent Coverage. Int. J. Mol. Sci. 2014, 15, 4189-4200. https://doi.org/10.3390/ijms15034189
Liu C, Bai C, Guo Y, Liu D, Lu T, Li X, Ma J, Ma Y, Guan W. Construction and Analysis of Siberian Tiger Bacterial Artificial Chromosome Library with Approximately 6.5-Fold Genome Equivalent Coverage. International Journal of Molecular Sciences. 2014; 15(3):4189-4200. https://doi.org/10.3390/ijms15034189
Chicago/Turabian StyleLiu, Changqing, Chunyu Bai, Yu Guo, Dan Liu, Taofeng Lu, Xiangchen Li, Jianzhang Ma, Yuehui Ma, and Weijun Guan. 2014. "Construction and Analysis of Siberian Tiger Bacterial Artificial Chromosome Library with Approximately 6.5-Fold Genome Equivalent Coverage" International Journal of Molecular Sciences 15, no. 3: 4189-4200. https://doi.org/10.3390/ijms15034189




