Risk-Association of Five SNPs in TOX3/LOC643714 with Breast Cancer in Southern China
Abstract
:1. Introduction
2. Results and Discussion
2.1. Subject Characteristics
2.2. Associations between Five SNPs and Breast Cancer Risk
2.3. Combined Effect of SNP rs4784227 and rs8051542 in TOX3/LOC643714
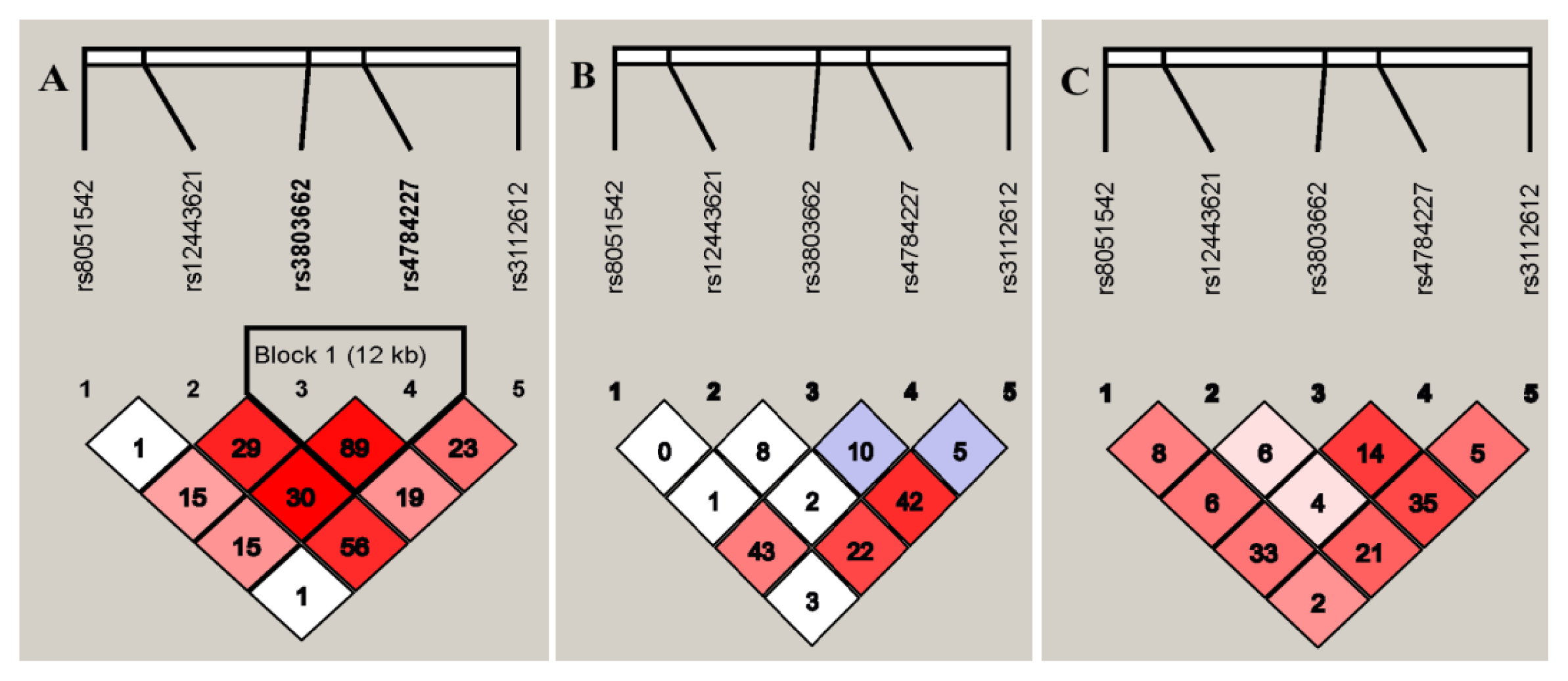
2.4. Linkage Disequilibrium of the SNPs in TOX3/LOC643714
3. Experimental Section
3.1. Subjects
3.2. DNA Extraction
3.3. SNP Selection and Genotyping
3.4. Statistical Analysis
4. Conclusions
Supplementary Information
ijms-15-02130-s001.pdfAcknowledgments
Conflicts of Interest
References
- Ferlay, J.; Shin, H.R.; Bray, F.; Forman, D.; Mathers, C.; Parkin, D.M. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int. J. Cancer 2010, 127, 2893–2917. [Google Scholar]
- Yang, L.; Parkin, D.M.; Li, L.D.; Chen, Y.D.; Bray, F. Estimation and projection of the national profile of cancer mortality in China: 1991–2005. Br. J. Cancer 2004, 90, 2157–2166. [Google Scholar]
- Dapic, V.; Carvalho, M.A.; Monteiro, A.N. Breast cancer susceptibility and the DNA damage response. Cancer Control 2005, 12, 127–136. [Google Scholar]
- Walsh, T.; King, M.C. Ten genes for inherited breast cancer. Cancer Cell 2007, 11, 103–105. [Google Scholar]
- Pharoah, P.D.; Dunning, A.M.; Ponder, B.A.; Easton, D.F. Association studies for finding cancer-susceptibility genetic variants. Nat. Rev. Cancer 2004, 4, 850–860. [Google Scholar]
- Easton, D.F.; Pooley, K.A.; Dunning, A.M.; Pharoah, P.D.; Thompson, D.; Ballinger, D.G.; Struewing, J.P.; Morrison, J.; Field, H.; Luben, R.; et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature 2007, 447, 1087–1093. [Google Scholar]
- Stacey, S.N.; Manolescu, A.; Sulem, P.; Rafnar, T.; Gudmundsson, J.; Gudjonsson, S.A.; Masson, G.; Jakobsdottir, M.; Thorlacius, S.; Helgason, A.; et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat. Genet 2007, 39, 865–869. [Google Scholar]
- Hunter, D.J.; Kraft, P.; Jacobs, K.B.; Cox, D.G.; Yeager, M.; Hankinson, S.E.; Wacholder, S.; Wang, Z.; Welch, R.; Hutchinson, A.; et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet 2007, 39, 870–874. [Google Scholar]
- Cox, A.; Dunning, A.M.; Garcia-Closas, M.; Balasubramanian, S.; Reed, M.W.; Pooley, K.A.; Scollen, S.; Baynes, C.; Ponder, B.A.; Chanock, S.; et al. A common coding variant in CASP8 is associated with breast cancer risk. Nat. Genet 2007, 39, 352–358. [Google Scholar]
- Thomas, G.; Jacobs, K.B.; Kraft, P.; Yeager, M.; Wacholder, S.; Cox, D.G.; Hankinson, S.E.; Hutchinson, A.; Wang, Z.; Yu, K.; et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1). Nat. Genet 2009, 41, 579–584. [Google Scholar]
- Reeves, G.K.; Travis, R.C.; Green, J.; Bull, D.; Tipper, S.; Baker, K.; Beral, V.; Peto, R.; Bell, J.; Zelenika, D.; et al. Incidence of breast cancer and its subtypes in relation to individual and multiple low-penetrance genetic susceptibility loci. JAMA 2010, 304, 426–434. [Google Scholar]
- Ruiz-Narvaez, E.A.; Rosenberg, L.; Cozier, Y.C.; Cupples, L.A.; Adams-Campbell, L.L.; Palmer, J.R. Polymorphisms in the TOX3/LOC643714 locus and risk of breast cancer in African-American women. Cancer Epidemiol. Biomark. Prev 2010, 19, 1320–1327. [Google Scholar]
- Long, J.; Shu, X.O.; Cai, Q.; Gao, Y.T.; Zheng, Y.; Li, G.; Li, C.; Gu, K.; Wen, W.; Xiang, Y.B.; et al. Evaluation of breast cancer susceptibility loci in Chinese women. Cancer Epidemiol. Biomark. Prev 2010, 19, 2357–2365. [Google Scholar]
- Zheng, W.; Long, J.; Gao, Y.T.; Li, C.; Zheng, Y.; Xiang, Y.B.; Wen, W.; Levy, S.; Deming, S.L.; Haines, J.L.; et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat. Genet 2009, 41, 324–328. [Google Scholar]
- Zheng, W.; Wen, W.; Gao, Y.T.; Shyr, Y.; Zheng, Y.; Long, J.; Li, G.; Li, C.; Gu, K.; Cai, Q.; et al. Genetic and clinical predictors for breast cancer risk assessment and stratification among Chinese women. J. Natl. Cancer Inst 2010, 102, 972–981. [Google Scholar]
- Greenman, C.; Stephens, P.; Smith, R.; Dalgliesh, G.L.; Hunter, C.; Bignell, G.; Davies, H.; Teague, J.; Butler, A.; Stevens, C.; et al. Patterns of somatic mutation in human cancer genomes. Nature 2007, 446, 153–158. [Google Scholar]
- Yuan, S.H.; Qiu, Z.; Ghosh, A. TOX3 regulates calcium-dependent transcription in neurons. Proc. Natl. Acad. Sci. USA 2009, 106, 2909–2914. [Google Scholar]
- Smid, M.; Wang, Y.; Klijn, J.G.; Sieuwerts, A.M.; Zhang, Y.; Atkins, D.; Martens, J.W.; Foekens, J.A. Genes associated with breast cancer metastatic to bone. J. Clin. Oncol 2006, 24, 2261–2267. [Google Scholar]
- Long, J.; Cai, Q.; Shu, X.O.; Qu, S.; Li, C.; Zheng, Y.; Gu, K.; Wang, W.; Xiang, Y.B.; Cheng, J.; et al. Identification of a functional genetic variant at 16q12.1 for breast cancer risk: Results from the Asia Breast Cancer Consortium. PLoS Genet 2010, 6, e1001002. [Google Scholar]
- Udler, M.S.; Ahmed, S.; Healey, C.S.; Meyer, K.; Struewing, J.; Maranian, M.; Kwon, E.M.; Zhang, J.; Tyrer, J.; Karlins, E.; et al. Fine scale mapping of the breast cancer 16q12 locus. Hum. Mol. Genet 2010, 19, 2507–2515. [Google Scholar]
- National center for biotechnology information. Available online: http://www.ncbi.nlm.nih.gov/gene/643714 (accessed on 7 January 2014).
- Travers, A.A. Priming the nucleosome: A role for HMGB proteins? EMBO Rep 2003, 4, 131–136. [Google Scholar]
- Dittmer, S.; Kovacs, Z.; Yuan, S.H.; Siszler, G.; Kogl, M.; Summer, H.; Geerts, A.; Golz, S.; Shioda, T.; Methner, A. TOX3 is a neuronal survival factor that induces transcription depending on the presence of CITED1 or phosphorylated CREB in the transcriptionally active complex. J. Cell Sci 2011, 124, 252–260. [Google Scholar]
- Yahata, T.; Shao, W.; Endoh, H.; Hur, J.; Coser, K.R.; Sun, H.; Ueda, Y.; Kato, S.; Isselbacher, K.J.; Brown, M.; et al. Selective coactivation of estrogen-dependent transcription by CITED1 CBP/p300-binding protein. Genes Dev 2001, 15, 2598–2612. [Google Scholar]
- Shioda, T.; Lechleider, R.J.; Dunwoodie, S.L.; Li, H.; Yahata, T.; de Caestecker, M.P.; Fenner, M.H.; Roberts, A.B.; Isselbacher, K.J. Transcriptional activating activity of Smad4: Roles of SMAD hetero-oligomerization and enhancement by an associating transactivator. Proc. Natl. Acad. Sci. USA 1998, 95, 9785–9790. [Google Scholar]
- Kim, H.C.; Lee, J.Y.; Sung, H.; Choi, J.Y.; Park, S.K.; Lee, K.M.; Kim, Y.J.; Go, M.J.; Li, L.; Cho, Y.S.; et al. A genome-wide association study identifies a breast cancer risk variant in ERBB4 at 2q34: Results from the Seoul Breast Cancer Study. Breast Cancer Res 2012, 14, R56. [Google Scholar]
- Sueta, A.; Ito, H.; Kawase, T.; Hirose, K.; Hosono, S.; Yatabe, Y.; Tajima, K.; Tanaka, H.; Iwata, H.; Iwase, H.; et al. A genetic risk predictor for breast cancer using a combination of low-penetrance polymorphisms in a Japanese population. Breast Cancer Res. Treat 2012, 132, 711–721. [Google Scholar]
- Garcia-Closas, M.; Hall, P.; Nevanlinna, H.; Pooley, K.; Morrison, J.; Richesson, D.A.; Bojesen, S.E.; Nordestgaard, B.G.; Axelsson, C.K.; Arias, J.I.; et al. Heterogeneity of breast cancer associations with five susceptibility loci by clinical and pathological characteristics. PLoS Genet 2008, 4, e1000054. [Google Scholar]
- Li, L.; Zhou, X.; Huang, Z.; Liu, Z.; Song, M.; Guo, Z. TNRC9/LOC643714 polymorphisms are not associated with breast cancer risk in Chinese women. Eur. J. Cancer Prev 2009, 18, 285–290. [Google Scholar]
- Liang, J.; Chen, P.; Hu, Z.; Shen, H.; Wang, F.; Chen, L.; Li, M.; Tang, J.; Wang, H. Genetic variants in trinucleotide repeat-containing 9 (TNRC9) are associated with risk of estrogen receptor positive breast cancer in a Chinese population. Breast Cancer Res. Treat 2010, 124, 237–241. [Google Scholar]
- Rinella, E.S.; Shao, Y.; Yackowski, L.; Pramanik, S.; Oratz, R.; Schnabel, F.; Guha, S.; Leduc, C.; Campbell, C.L.; Klugman, S.D.; et al. Genetic variants associated with breast cancer risk for Ashkenazi Jewish women with strong family histories but no identifiable BRCA1/2 mutation. Hum. Genet 2013, 132, 523–536. [Google Scholar]
- Cowper-Sallari, R.; Zhang, X.; Wright, J.B.; Bailey, S.D.; Cole, M.D.; Eeckhoute, J.; Moore, J.H.; Lupien, M. Breast cancer risk-associated SNPs modulate the affinity of chromatin for FOXA1 and alter gene expression. Nat. Genet 2012, 44, 1191–1198. [Google Scholar]
- Sole, X.; Guino, E.; Valls, J.; Iniesta, R.; Moreno, V. SNPStats: A web tool for the analysis of association studies. Bioinformatics 2006, 22, 1928–1929. [Google Scholar]
| Variable | Value |
|---|---|
| Age, years (mean ± SD) | 48.5 ± 10.0 (range 22–80) |
| Tumor histology (n = 559) | |
| IDC | 524 (84.1%) |
| Others a | 35 (5.6%) |
| Unknown | 64 (10.3%) |
| Clinical staging of cancer (UICC) b (n = 559) | |
| Stage 0 (in situ) | 6 (1.0%) |
| Stage 1 | 153 (24.6%) |
| Stage 2 | 154 (24.7%) |
| Stage 3 | 187 (30.0%) |
| Stage 4 | 59 (9.5%) |
| Unknown | 64 (10.3%) |
| Receptor status | |
| Estrogen receptor (n = 517) | |
| Positive | 300 (48.2%) |
| Negative | 217 (34.8%) |
| Unknown | 106 (17.0%) |
| Progesterone receptor (n = 516) | |
| Positive | 267 (42.9%) |
| Negative | 249 (40.0%) |
| Unknown | 107 (17.2%) |
| Human epidermal growth factor receptor 2 (n = 490) | |
| Positive | 230 (36.9%) |
| Negative | 260 (41.7%) |
| Unknown | 133 (21.3%) |
| Triple-negative c | 74 (11.9%) |
| Luminal A d | 185 (29.7%) |
| SNP | Position | Alleles (reference/risk) | MAF (control/case) | Codominant model | Additive model | ||
|---|---|---|---|---|---|---|---|
| Heterozygote OR (95% CI) a | Homozygote OR (95% CI) a | Per-allele OR (95% CI) a | Ptrend b | ||||
| rs8051542 | 52534167 | C/T | 0.17/0.20 | 1.26 (0.98–1.62) | 1.60 (0.83–3.10) | 1.26 (1.02–1.56) | 0.030 |
| rs12443621 | 52548037 | A/G | 0.43/0.42 | 1.02 (0.74–1.40) | 1.04 (0.74–1.45) | 1.02 (0.86–1.20) | 0.827 |
| rs3803662 | 52586341 | C/T | 0.66/0.66 | 1.10 (0.76–1.59) | 1.07 (0.74–1.56) | 1.02 (0.86–1.21) | 0.826 |
| rs4784227 | 52599188 | C/T | 0.24/0.30 | 1.42 (1.12–1.81) | 1.51 (0.97–2.35) | 1.31 (1.10–1.57) | 0.003 |
| rs3112612 | 52635164 | T/C | 0.21/0.21 | 0.91 (0.71–1.16) | 1.18 (0.68–2.04) | 0.98 (0.81–1.19) | 0.850 |
| Subtypes | T-rs8051542 | T-rs4784227 | ||||
|---|---|---|---|---|---|---|
| Cases | OR (95% CI) a | p value | Cases | OR (95% CI) a | p value | |
| Estrogen receptor (ER) | ||||||
| ER-positive | 596 | 1.31 (1.02–1.69) | 0.034 | 598 | 1.41 (1.13–1.75) | 0.002 |
| ER-negative | 430 | 1.18 (0.89–1.58) | 0.246 | 434 | 1.42 (1.11–1.81) | 0.005 |
| Progesterone receptor (PR) | ||||||
| PR-positive | 530 | 1.40 (1.08–1.81) | 0.100 | 534 | 1.43 (1.14–1.79) | 0.002 |
| PR-negative | 494 | 1.10 (0.83–1.46) | 0.486 | 484 | 1.39 (1.10–1.75) | 0.007 |
| Human epidermal growth factor receptor 2 (HER2) | ||||||
| HER2-positive | 456 | 1.24 (0.94–1.64) | 0.131 | 458 | 1.37 (1.08–1.75) | 0.010 |
| HER2-negative | 516 | 1.30 (1.00–1.69) | 0.051 | 520 | 1.51 (1.20–1.89) | 0.000 |
| Dominant model | rs8051542 | Additive model | rs8051542 | ||
|---|---|---|---|---|---|
| rs4784227 | CC | CTTT | rs4784227 | C | T |
| CC | 1.00 | 1.03 (0.59–1.77) | C | 1.00 | 1.09 (0.68–1.73) |
| CTTT | 1.36 (0.99–1.87) | 1.49 (1.14–1.95) | T | 1.29 (0.99–1.66) | 1.37 (1.09–1.72) |
© 2014 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
He, X.; Yao, G.; Li, F.; Li, M.; Yang, X. Risk-Association of Five SNPs in TOX3/LOC643714 with Breast Cancer in Southern China. Int. J. Mol. Sci. 2014, 15, 2130-2141. https://doi.org/10.3390/ijms15022130
He X, Yao G, Li F, Li M, Yang X. Risk-Association of Five SNPs in TOX3/LOC643714 with Breast Cancer in Southern China. International Journal of Molecular Sciences. 2014; 15(2):2130-2141. https://doi.org/10.3390/ijms15022130
Chicago/Turabian StyleHe, Xuanqiu, Guangyu Yao, Fenxia Li, Ming Li, and Xuexi Yang. 2014. "Risk-Association of Five SNPs in TOX3/LOC643714 with Breast Cancer in Southern China" International Journal of Molecular Sciences 15, no. 2: 2130-2141. https://doi.org/10.3390/ijms15022130
APA StyleHe, X., Yao, G., Li, F., Li, M., & Yang, X. (2014). Risk-Association of Five SNPs in TOX3/LOC643714 with Breast Cancer in Southern China. International Journal of Molecular Sciences, 15(2), 2130-2141. https://doi.org/10.3390/ijms15022130




