Cloning and Characterization of Farnesyl Diphosphate Synthase Gene Involved in Triterpenoids Biosynthesis from Poria cocos
Abstract
:1. Introduction
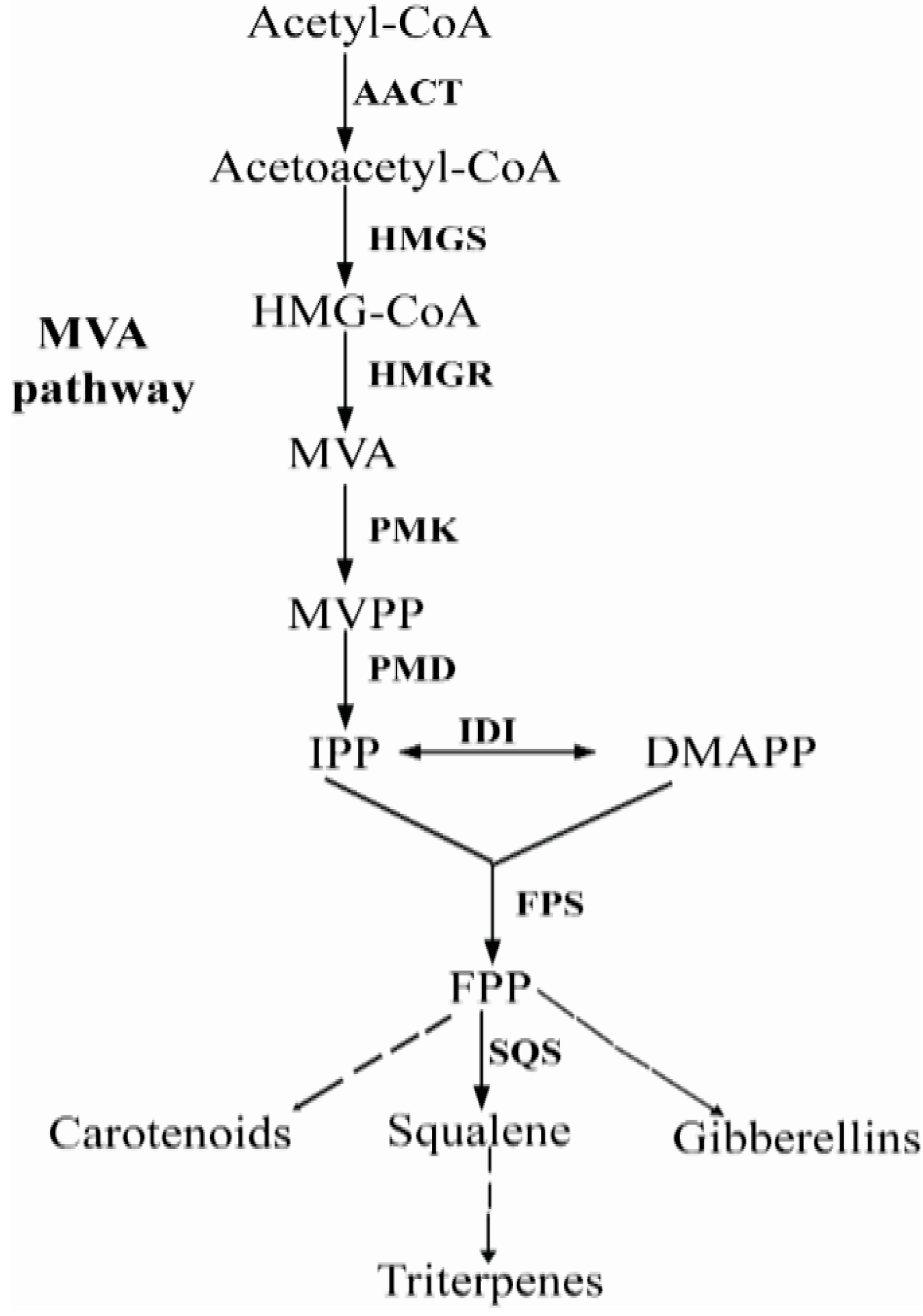
2. Results and Discussion
2.1. Isolation and Sequence Analysis of Pc-fps
2.2. Sequence Features of the Pc-fps Promoter Region

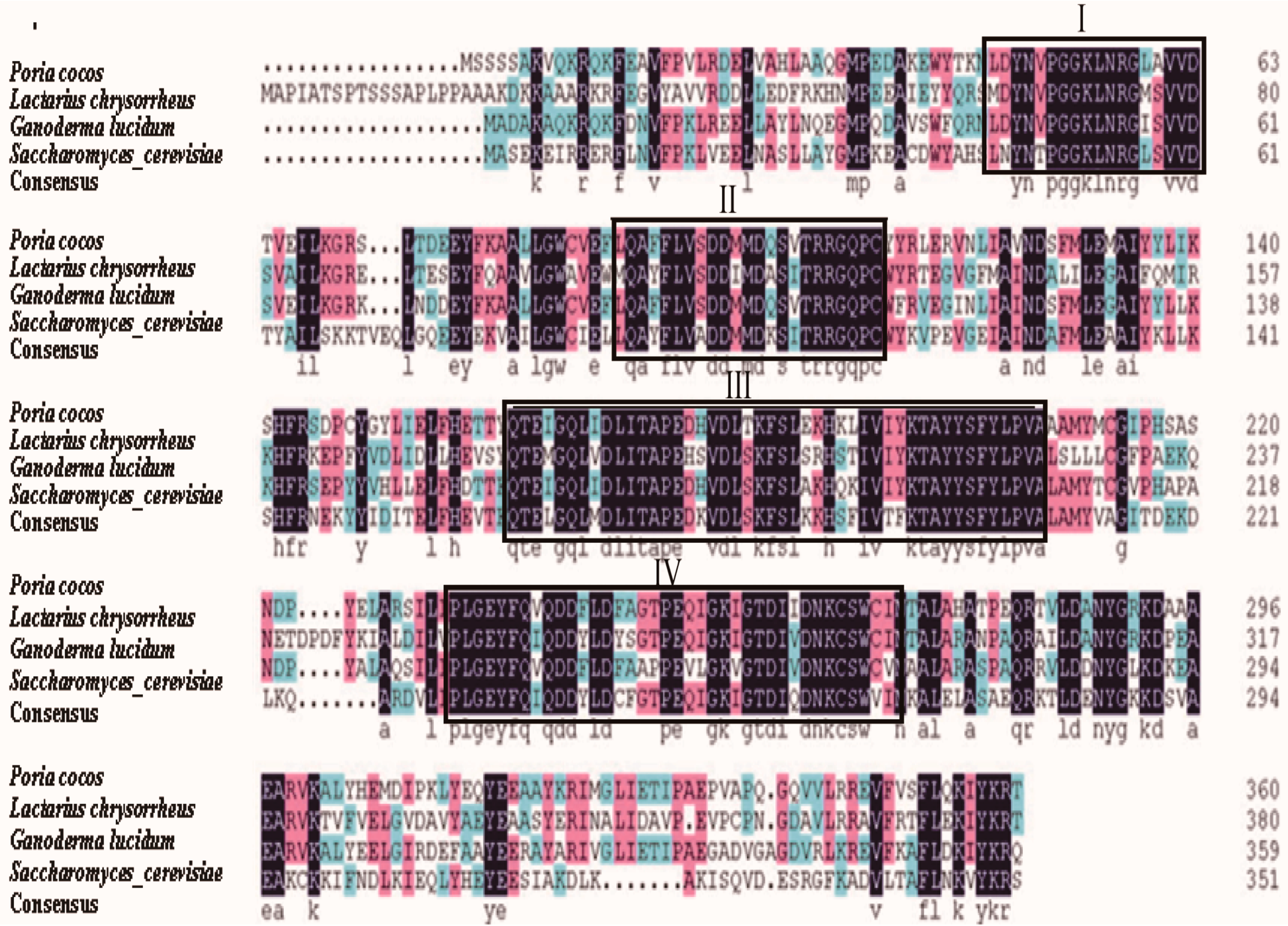
2.3. Comparison of FPS Protein Sequences
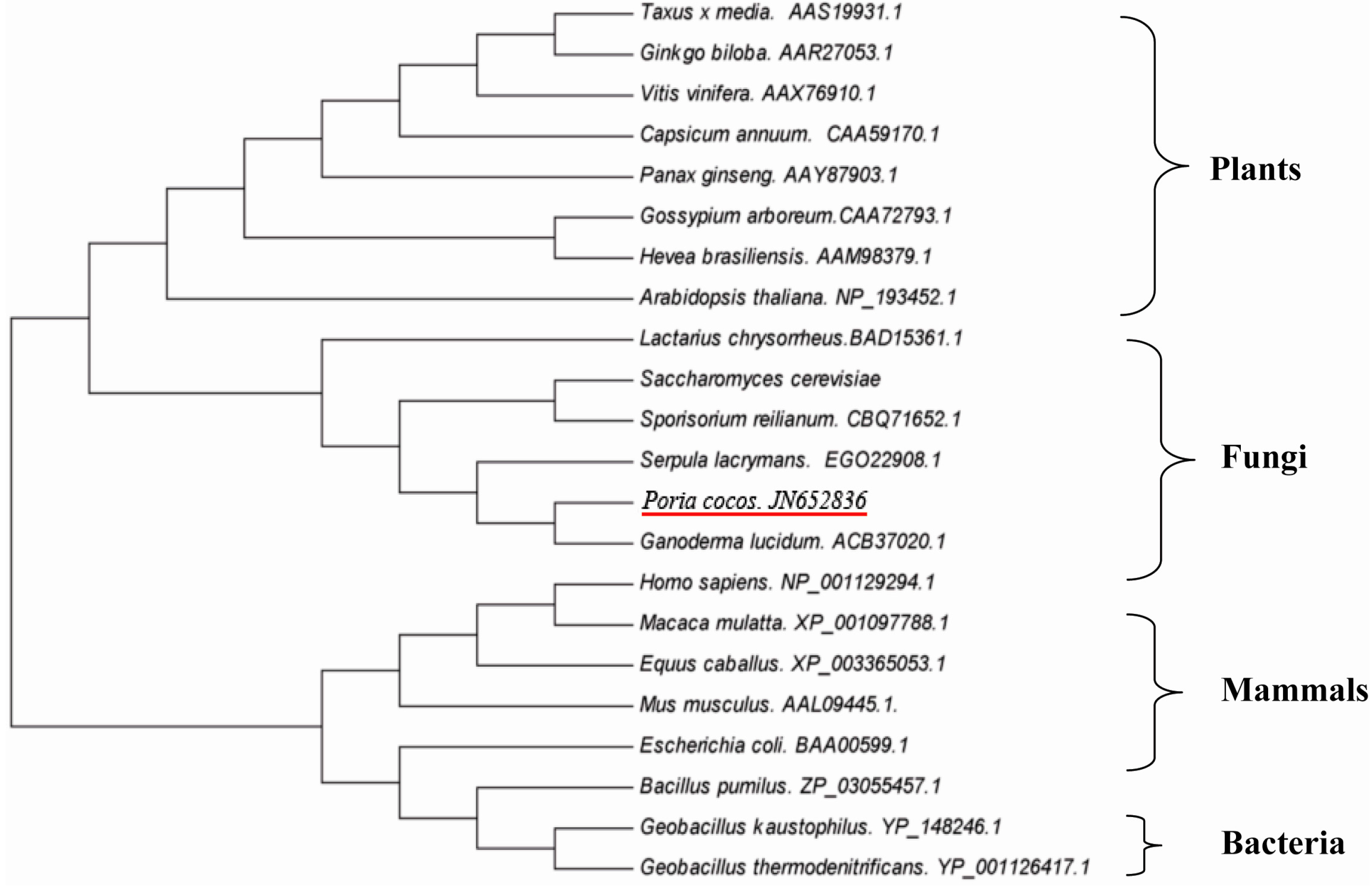
2.4. Phylogenetic Analysis
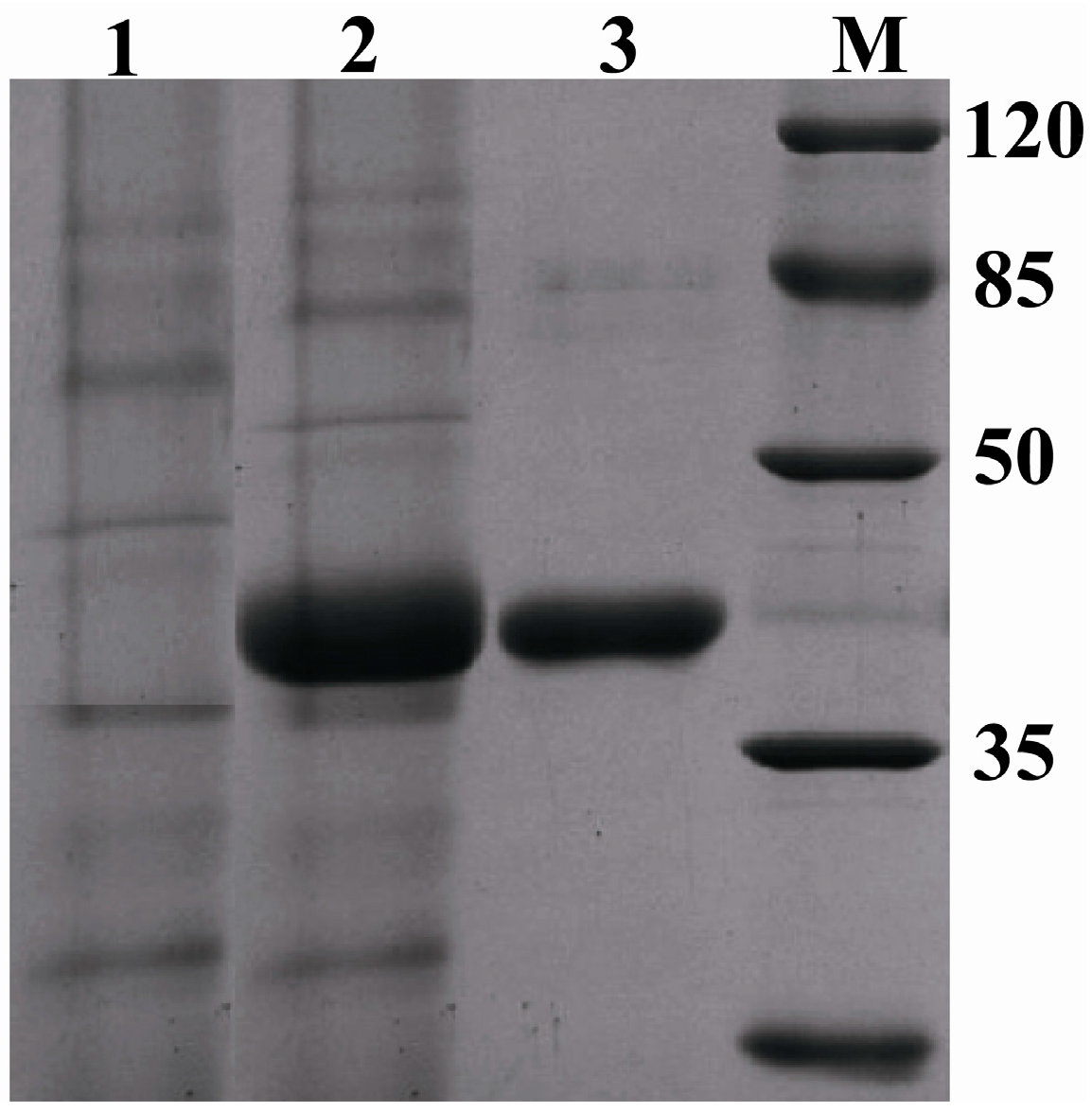
2.5. Expression of Pc-FPS in Pichia pastoris (P. pastoris)
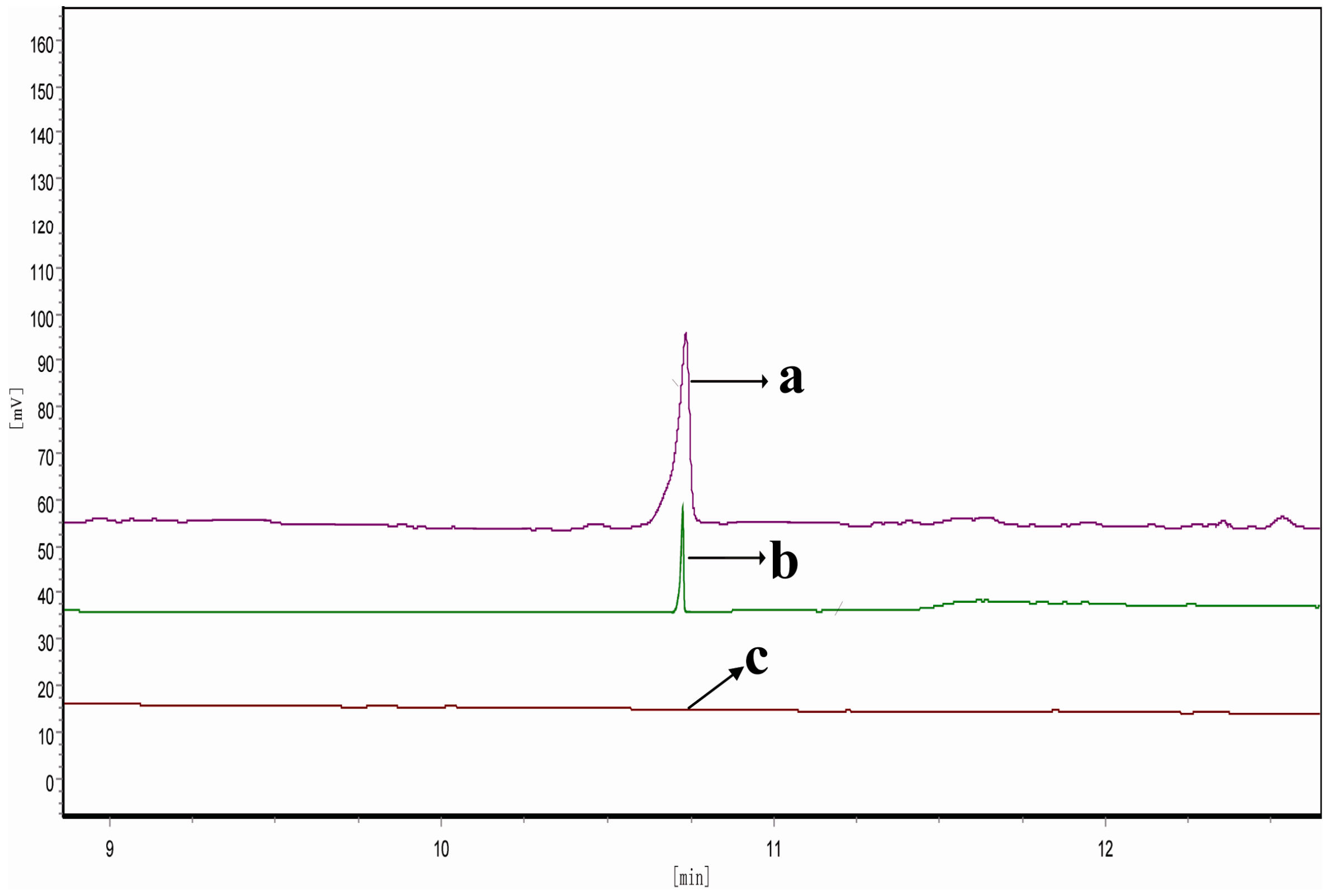
2.6. Determination of FPP-Derived Farnesol by Gas Chromatography (GC)
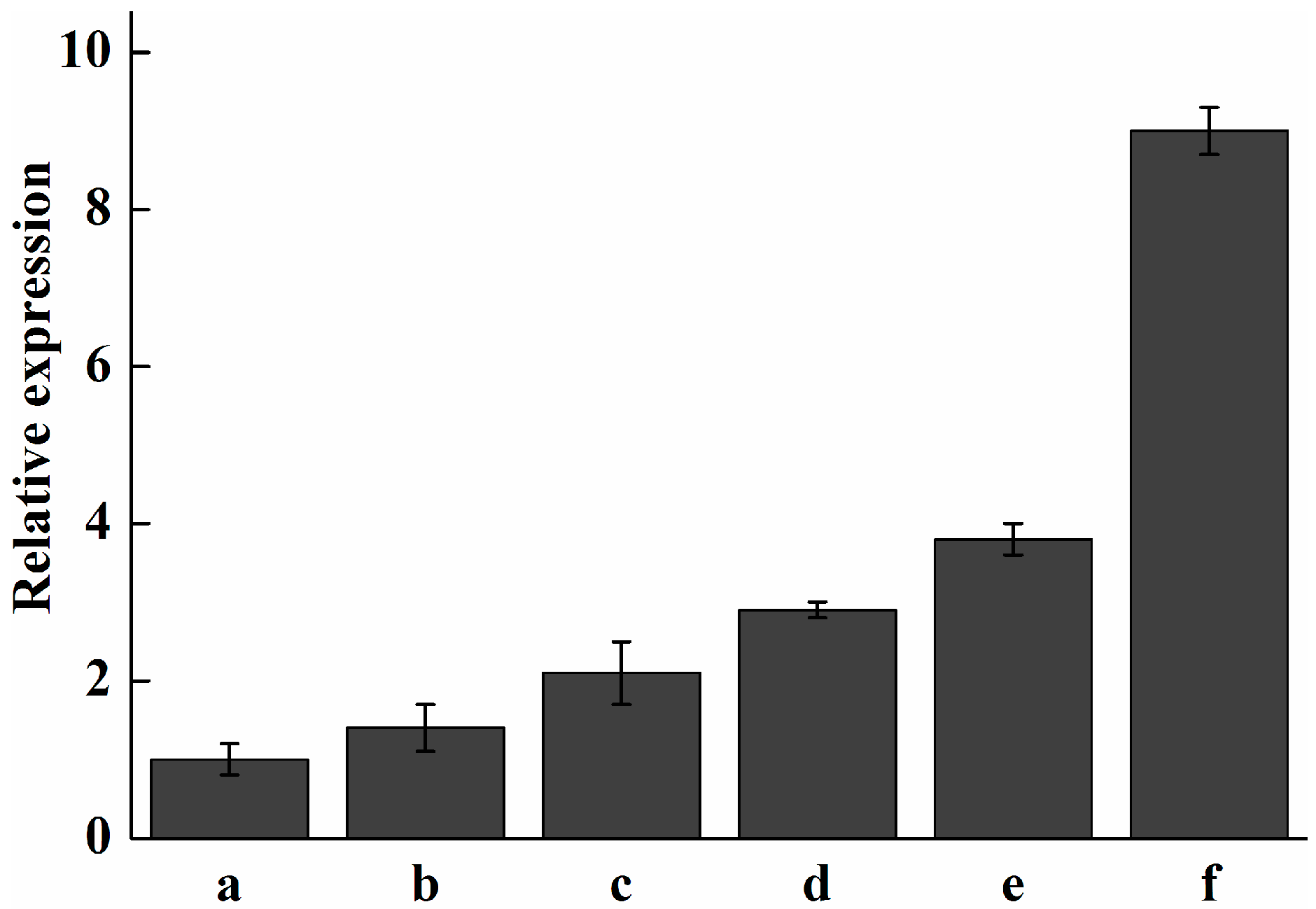
2.7. Analysis of Expression Profile of Pc-fps and Total Triterpenoids at Different Stages of Development

2.8. Analysis of Expression Profile of Pc-fps and Content of Triterpenoids under Methyl Jasmonate (MeJA) Treatments
3. Experimental Section
3.1. Cultures and Materials

3.2. Cloning of Pc-fps
3.3. Bioinformatics Analysis
3.4. Vector Construction and Transformation and Selection of P. pastoris Clones
3.5. Shaking Flask Cultures and Purification of the Recombinant FPS (rFPS)
3.6. Determination of Enzymatic Activity and Product Analysis by GC
3.7. Expression Profile Analysis of Pc-fps under Different Stages of Development
3.8. Extraction and Analysis of Total Triterpenoids under Different Stages of Development
3.9. Analysis of Expression Profile of Pc-fps and Content of Triterpenoids under Methyl Jasmonate (MeJA) Treatments
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Cuella, M.J.; Giner, R.M.; Recio, M.C.; Just, M.J.; Manez, S.; Rios, J.L. Two fungal lanostane derivatives as phospholipase A2 inhibitors. J. Nat. Prod. 1996, 59, 977–979. [Google Scholar] [CrossRef]
- Tai, T.; Akita, Y.; Kinoshita, K.; Koyama, K.; Takahashi, K.; Watanabe, K. Anti-emetic principles of Poria cocos. Planta Med. 1995, 61, 527–530. [Google Scholar] [CrossRef]
- Kikuchi, T.; Uchiyama, E.; Ukiya, M.; Tabata, K.; Kimura, Y.; Suzuki, T.; Akihisa, T. Cytotoxic and apoptosis inducing activities of triterpene acids from Poria cocos. J. Nat. Prod. 2011, 74, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Akihisa, T.; Nakamura, Y.; Tokuda, H.; Uchiyama, E.; Suzuki, T.; Kimura, Y.; Uchikura, K.; Nishino, H. Triterpene acids from Poria cocos and their antitumor promoting effects. J. Nat. Prod. 2007, 70, 948–953. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K. Non-mevalonate isoprenoid biosynthesis: enzymes, genes and inhibitors. Biochem. Soc. Trans. 2000, 28, 785–789. [Google Scholar] [CrossRef] [PubMed]
- Lombard, J.; Moreira, D. Origins and early evolution of the mevalonate pathway of isoprenoid biosynthesis in the three domains of life. Mol. Biol. Evol. 2011, 28, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Hunter, W.N. The non-mevalonate pathway of isoprenoid precursor biosynthesis. J. Biol. Chem. 2007, 282, 21573–21577. [Google Scholar] [CrossRef] [PubMed]
- Withers, S.T.; Keasling, J.D. Biosynthesis and engineering of isoprenoid small molecules. Appl. Microbiol. Biotechnol. 2007, 73, 980–990. [Google Scholar] [CrossRef] [PubMed]
- Lange, B.M.; Rujan, T.; Martin, W.; Croteau, R. Isoprenoid biosynthesis: the evolution of two ancient and distinct pathways across genomes. Proc. Natl. Acad. Sci. USA 2000, 97, 13172–13177. [Google Scholar] [CrossRef] [PubMed]
- Delourme, D.; Lacroute, F.; Karst, F. Cloning of an Arabidopsis thaliana cDNA coding for farnesyl diphosphate synthase by functional complementation in yeast. Plant Mol. Biol. 1994, 26, 1867–1873. [Google Scholar] [CrossRef]
- Dhar, M.K.; Koul, A.; Kaul, S. Farnesyl pyrophosphate synthase: a key enzyme in isoprenoid biosynthetic pathway and potential molecular target for drug development. New Biotechnol. 2013, 30, 114–123. [Google Scholar] [CrossRef]
- Ding, Y.X.; Yang, X.O.; Shang, C.H.; Ren, A.; Shi, L.; Li, Y.X.; Zhao, M.W. Molecular cloning, characterization, and differential expression of a farnesyl diphosphate synthase gene from the basidiomycetous fungus Ganoderma lucidum. Biosci. Biotechnol. Biochem. 2008, 72, 1571–1579. [Google Scholar] [CrossRef]
- Keim, V.; Manzano, D.; Fernández, F.J.; Closa, M.; Andrade, P.; Caudepón, D.; Bortolotti, C.; Vega, M.C.; Arró, M.; Ferrer, A. Characterization of Arabidopsis FPS isozymes and FPS gene expression analysis provide insight into the biosynthesis of isoprenoid precursors in seeds. PLoS One 2012, 7, e49109. [Google Scholar] [CrossRef] [Green Version]
- Cervantes-Cervantes, M.; Gallagher, C.E.; Zhu, C.; Wurtzel, E.T. Maize cDNAs expressed in endosperm encode functional farnesyl diphosphate synthase with geranylgeranyl diphosphate synthase activity. Plant Physiol. 2006, 141, 220–231. [Google Scholar] [CrossRef] [PubMed]
- Hemmerlin, A.; Rivera, S.B.; Erickson, H.K.; Poulter, C.D. Enzymes encoded by the farnesyl diphosphate synthase gene family in the Big Sagebrush Artemisia tridentata ssp. spiciformis. J. Biol. Chem. 2003, 278, 32132–32140. [Google Scholar] [CrossRef] [PubMed]
- Romanelli, M.G.; Lorenzi, P.; Sangalli, A.; Diani, E.; Mottes, M. Characterization and functional analysis of cis-acting elements of the human farnesyl diphosphate synthetase (FDPS) gene 5' flanking region. Genomics 2009, 93, 227–234. [Google Scholar] [CrossRef]
- Guan, G.; Jiang, G.; Koch, R.L.; Shechter, I. Molecular cloning and functional analysis of the promoter of the human squalene synthase gene. J. Biol. Chem. 1995, 270, 21958–21965. [Google Scholar] [CrossRef] [PubMed]
- Ashby, M.N.; Edwards, P.A. Elucidation of the deficiency in two yeast coenzyme Q mutants. Characterization of the structural gene encoding hexaprenyl pyrophosphate synthetase. J. Biol. Chem. 1990, 265, 13157–13164. [Google Scholar] [PubMed]
- Tarshis, L.C.; Proteau, P.J.; Kellogg, B.A.; Sacchettini, J.C.; Poulter, C.D. Regulation of product chain length by isoprenyl diphosphate synthases. Proc. Natl. Acad. Sci. USA 1996, 93, 15018–15023. [Google Scholar] [CrossRef] [PubMed]
- Blanchard, L.; Karst, F. Characterization of a lysine-to-glutamic acid mutation in a conservative sequence of farnesyl diphosphate synthase from Saccharomyces cerevisiae. Gene 1993, 125, 185–189. [Google Scholar] [CrossRef] [PubMed]
- Liechti, R.; Farmer, E.E. The jasmonate pathway. Science 2002, 296, 1649–1650. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.R.; Lin, J.F.; Guo, L.Q.; You, L.F.; Zeng, X.L.; Wen, J.M. Cloning and characterization of squalene synthase gene from Poria. cocos and its up-regulation by methyl jasmonate. World J. Microbiol. Biotechnol. 2014, 30, 613–620. [Google Scholar] [CrossRef]
- Wang, S.Y.; Bowman, L.; Ding, M. Methyl jasmonate enhances antioxidant activity and flavonoid content in blackberries (Rubus. sp.) and promotes antiproliferation of human cancer cells. Food Chem. 2008, 107, 1261–1269. [Google Scholar]
- Ren, A.; Qin, L.; Shi, L.; Dong, X.; Mu, D.S.; Li, Y.X.; Zhao, M.W. Methyl jasmonate induces ganoderic acid biosynthesis in the basidiomycetous fungus Ganoderma lucidum. Bioresour. Technol. 2010, 101, 6785–6490. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef] [PubMed]
- Mekkriengkrai, D.; Sando, T.; Hirooka, K.; Sakdapipanich, J.; Tanaka, Y.; Fukusaki, E.; Kobayashi, A. Cloning and characterization of farnesyl diphosphate synthase from the rubber-producing mushroom Lactarius chrysorrheus. Biosci. Biotechnol. Biochem. 2004, 68, 2360–2368. [Google Scholar] [CrossRef]
- Zhao, M.W.; Liang, W.Q.; Zhang, D.B.; Wang, N.; Wang, C.G.; Pan, Y.J. Cloning and characterization of squalene synthase (SQS) gene from Ganoderma lucidum. J. Microbiol. Biotechnol. 2007, 17, 1106–1112. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Shu, S.; Chen, B.; Zhou, M.; Zhao, X.; Xia, H.; Wang, M. De novo sequencing and transcriptome analysis of Wolfiporia cocos to reveal genes related to biosynthesis of triterpenoids. PLoS One 2013, 14, e71350. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Li, Y.; Liu, D. Cloning and Characterization of Farnesyl Diphosphate Synthase Gene Involved in Triterpenoids Biosynthesis from Poria cocos. Int. J. Mol. Sci. 2014, 15, 22188-22202. https://doi.org/10.3390/ijms151222188
Wang J, Li Y, Liu D. Cloning and Characterization of Farnesyl Diphosphate Synthase Gene Involved in Triterpenoids Biosynthesis from Poria cocos. International Journal of Molecular Sciences. 2014; 15(12):22188-22202. https://doi.org/10.3390/ijms151222188
Chicago/Turabian StyleWang, Jianrong, Yangyuan Li, and Danni Liu. 2014. "Cloning and Characterization of Farnesyl Diphosphate Synthase Gene Involved in Triterpenoids Biosynthesis from Poria cocos" International Journal of Molecular Sciences 15, no. 12: 22188-22202. https://doi.org/10.3390/ijms151222188



