Identification and Evaluation of Reliable Reference Genes for Quantitative Real-Time PCR Analysis in Tea Plant (Camellia sinensis (L.) O. Kuntze)
Abstract
:1. Introduction
2. Results
| Reference Gene | GeneBank Accession Number | Arabidopsis Ortholog | Arabidopsis Locus Description | Forward/Reverse Primer Sequence (5'–3') | Amplicon Size (bp) | qRT-PCR Efficiency (%) |
|---|---|---|---|---|---|---|
| Cs18S rRNA1 | AY563528.1 | At3g41768 | 18S ribosomal RNA | TGGCCTTCGGGATCGGAGTAAT/GCTTTCGCAGTTGTTCGTCTTTCAT | 108 | 101 |
| CsACTIN1 | KA280216.1 | At5g09810 | Actin 2/7 | TGGGCCAGAAAGATGCTTATGTAGG/ATGCCAGATCTTTTCCATGTCATCC | 118 | 103 |
| CsCLATHRIN1 | KA291473.1 | AT5G46630 | Clathrin adaptor complex subunit | TAGAGCGGGTAGTGGAGACCTCGTT/TACCAAAGCCGGCTCGTATGAGATT | 129 | 102 |
| CsEF1 | KA280301.1 | AT5G60390 | Elongation factor 1 alpha | TTGGACAAGCTCAAGGCTGAACG/ATGGCCAGGAGCATCAATGACAGT | 110 | 98 |
| CsGAPDH1 | KA295375.1 | AT1G13440 | Glyceraldehyde-3-phosphate dehydrogenase | TTTTTGGCCTTAGGAACCCAGAGG/GGGCAGCAGCCTTATCCTTATCAGT | 107 | 93 |
| CsSAND1 | KM057790 | AT2G28390 | SAND family protein | TCCAATTGCCCCCTTAATGACTCA/GTAAGGGCAGGCAAACACCAGGTA | 109 | 106 |
| CsTIP1 | KA283116.1 | AT4G34270 | TIP41-like protein | GGGTGCTTATGAGATTGAGGGACAC/ATATGCCGCAGAATCAGATGGGTAT | 148 | 101 |
| CsTUBULIN1 | DQ444294.1 | XM_002871860 | Tubulin alpha-3 | TGCCGCTAATAACTTTGCCAGAGG/GCCACCGCCAACAGCGTTAGTA | 141 | 92 |
| CsUBQ1 | HM003234.1 | AT4G05320 | Ubiquitin | GCCGGAAAACAGCTTGAAGATGG/AGGACGGCTCAATAATCCCACCAC | 111 | 98 |
| CsUBC1 | KA281185.1 | AT4G27960 | Ubiquitin-conjugating enzyme | TGCTGGTGGGGTTTTTCTTGTTACC/AAGGCATATGCTCCCATTGCTGTTT | 124 | 92 |
| CsPTB1 | GAAC01052498.1 | AT3G01150 | Polypyrimidine tract-binding protein | TGACCAAGCACACTCCACACTATCG/TGCCCCCTTATCATCATCCACAA | 107 | 95 |
2.1. Expression Profiles of Candidate Reference Genes
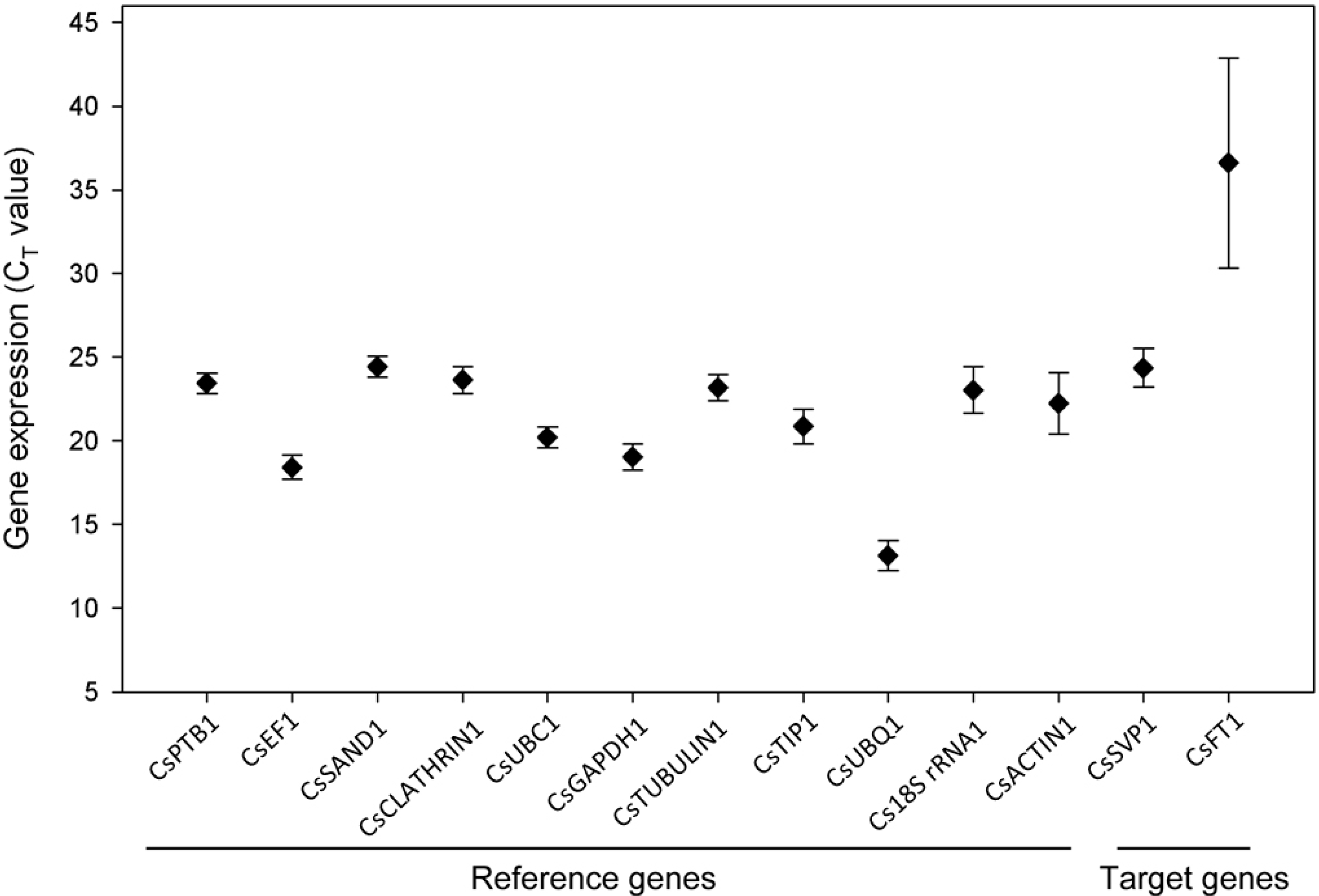

2.2. Stability Ranking of Candidate Reference Genes
3. Discussion
| Analysis Tool | Ranking Order (1 being the most stable, 11 being the least stable) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | |
| Diurnal Expression Ranking Order | |||||||||||
| ∆CT | CsEF1 | CsGAPDH1 | CsSAND1 | CsCLATHRIN1 | CsTIP1 | CsUBQ1 | CsUBC1 | CsPTB1 | Cs18S rRNA1 | CsACTIN1 | CsACTIN1 |
| BestKeeper | Cs18S rRNA1 | CsPTB1 | CsEF1 | CsSAND1 | CsTIP1 | CsGAPDH1 | CsUBC1 | CsCLATHRIN1 | CsACTIN1 | CsUBQ1 | CsACTIN1 |
| Normfinder | CsEF1 | CsGAPDH1 | CsUBQ1 | CsCLATHRIN1 | CsSAND1 | CsTIP1 | CsPTB1 | CsUBC1 | Cs18S rRNA1 | CsACTIN1 | CsACTIN1 |
| Genorm | CsGAPDH1|CsCLATHRIN1 | CsSAND1 | CsEF1 | CsTIP1 | CsUBC1 | CsUBQ1 | CsPTB1 | Cs18S rRNA1 | CsACTIN1 | CsACTIN1 | |
| Recommended comprehensive ranking | CsEF1 | CsGAPDH1 | CsCLATHRIN1 | CsSAND1 | Cs18S rRNA1 | CsTIP1 | CsPTB1 | CsUBQ1 | CsUBC1 | CsACTIN1 | CsACTIN1 |
| Different Organs Ranking Order | |||||||||||
| ∆CT | CsCLATHRIN1 | CsEF1 | CsGAPDH1 | CsPTB1 | CsSAND1 | CsTIP1 | CsUBC1 | CsUBQ1 | Cs18S rRNA1 | CsACTIN1 | CsACTIN1 |
| BestKeeper | Cs18S rRNA1 | CsPTB1 | CsSAND1 | CsTIP1 | CsCLATHRIN1 | CsEF1 | CsUBC1 | CsGAPDH1 | CsUBQ1 | CsACTIN1 | CsACTIN1 |
| Normfinder | CsCLATHRIN1 | CsUBC1 | CsGAPDH1 | CsEF1 | CsTIP1 | CsPTB1 | CsSAND1 | CsUBQ1 | Cs18S rRNA1 | CsACTIN1 | CsACTIN1 |
| Genorm | CsPTB1|CsSAND1 | CsEF1 | CsGAPDH1 | CsCLATHRIN1 | CsUBQ1 | CsTIP1 | CsUBC1 | Cs18S rRNA1 | CsACTIN1 | CsACTIN1 | |
| Recommended comprehensive ranking | CsCLATHRIN1 | CsPTB1 | CsSAND1 | CsEF1 | CsGAPDH1 | Cs18S rRNA1 | CsUBC1 | CsTIP1 | CsUBQ1 | CsACTIN1 | CsACTIN1 |
| Leaves with Cold and Short Photoperiod Treatment Ranking Order | |||||||||||
| ∆CT | CsCLATHRIN1 | CsGAPDH1 | CsPTB1 | CsUBQ1 | CsSAND1 | CsUBC1 | CsACTIN1 | CsEF1 | Cs18S rRNA1 | CsTIP1 | CsACTIN1 |
| BestKeeper | CsEF1 | CsSAND1 | Cs18S rRNA1 | CsGAPDH1 | CsUBC1 | CsPTB1 | CsCLATHRIN1 | CsACTIN1 | CsTIP1 | CsUBQ1 | CsACTIN1 |
| Normfinder | CsCLATHRIN1 | CsGAPDH1 | CsUBQ1 | CsPTB1 | CsSAND1 | CsUBC1 | CsACTIN1 | CsEF1 | Cs18S rRNA1 | CsTIP1 | CsACTIN1 |
| Genorm | CsCLATHRIN1|CsUBQ1 | CsGAPDH1 | CsPTB1 | CsUBC1 | CsSAND1 | CsEF1 | CsACTIN1 | Cs18S rRNA1 | CsTIP1 | CsACTIN1 | |
| Recommended comprehensive ranking | CsCLATHRIN1 | CsGAPDH1 | CsUBQ1 | CsPTB1 | CsSAND1 | CsEF1 | CsUBC1 | Cs18S rRNA1 | CsACTIN1 | CsTIP1 | CsACTIN1 |
| Shoots with Cold and Short Photoperiod Treatment Ranking Order | |||||||||||
| ∆CT | CsUBC1 | CsGAPDH1 | CsPTB1 | CsCLATHRIN1 | CsUBQ1 | CsEF1 | CsSAND1 | Cs18S rRNA1 | CsTIP1 | CsACTIN1 | CsACTIN1 |
| BestKeeper | CsPTB1 | CsGAPDH1 | CsUBC1 | CsCLATHRIN1 | CsEF1 | Cs18S rRNA1 | CsACTIN1 | CsSAND1 | CsTIP1 | CsUBQ1 | CsACTIN1 |
| Normfinder | CsUBC1 | CsGAPDH1 | CsPTB1 | CsCLATHRIN1 | CsUBQ1 | CsEF1 | Cs18S rRNA1 | CsSAND1 | CsTIP1 | CsACTIN1 | CsACTIN1 |
| Genorm | CsUBC1|CsCLATHRIN1 | CsPTB1 | CsTIP1 | CsSAND1 | CsGAPDH1 | CsUBQ1 | CsEF1 | Cs18S rRNA1 | CsACTIN1 | CsACTIN1 | |
| Recommended comprehensive ranking | CsUBC1 | CsPTB1 | CsGAPDH1 | CsCLATHRIN1 | CsEF1 | CsUBQ1 | CsSAND1 | CsTIP1 | Cs18S rRNA1 | CsACTIN1 | CsACTIN1 |
| Auxin Antagonist Auxinole Treatment Ranking Order | |||||||||||
| ∆CT | CsPTB1 | CsCLATHRIN1 | CsUBC1 | CsSAND1 | CsEF1 | CsACTIN1 | CsUBQ1 | CsTIP1 | CsGAPDH1 | CsACTIN1 | Cs18S rRNA1 |
| BestKeeper | CsPTB1 | CsUBC1 | CsCLATHRIN1 | CsACTIN1 | CsEF1 | Cs18S rRNA1 | CsSAND1 | CsACTIN1 | CsGAPDH1 | CsTIP1 | CsUBQ1 |
| Normfinder | CsPTB1 | CsCLATHRIN1 | CsUBC1 | CsSAND1 | CsEF1 | CsACTIN1 | CsUBQ1 | CsTIP1 | CsGAPDH1 | CsACTIN1 | Cs18S rRNA1 |
| Genorm | CsPTB1|CsCLATHRIN1 | CsUBC1 | CsACTIN1 | CsSAND1 | CsTIP1 | CsEF1 | CsUBQ1 | CsGAPDH1 | CsACTIN1 | Cs18S rRNA1 | |
| Recommended comprehensive ranking | CsPTB1 | CsCLATHRIN1 | CsUBC1 | CsSAND1 | CsACTIN1 | CsEF1 | CsTIP1 | CsUBQ1 | CsGAPDH1 | Cs18S rRNA1 | CsACTIN1 |
| Lanolin Treatment Ranking Order | |||||||||||
| ∆CT | CsPTB1 | CsUBC1 | CsCLATHRIN1 | CsEF1 | CsSAND1 | CsTIP1 | CsACTIN1 | CsGAPDH1 | Cs18S rRNA1 | CsUBQ1 | CsACTIN1 |
| BestKeeper | CsEF1 | CsUBC1 | CsPTB1 | CsCLATHRIN1 | CsSAND1 | CsTIP1 | CsACTIN1 | Cs18S rRNA1 | CsGAPDH1 | CsUBQ1 | CsACTIN1 |
| Normfinder | CsPTB1 | CsUBC1 | CsCLATHRIN1 | CsSAND1 | CsEF1 | CsTIP1 | CsACTIN1 | CsGAPDH1 | Cs18S rRNA1 | CsUBQ1 | CsACTIN1 |
| Genorm | CsUBC1|CsCLATHRIN1 | CsEF1 | CsPTB1 | CsTIP1 | CsSAND1 | CsACTIN1 | CsGAPDH1 | Cs18S rRNA1 | CsUBQ1 | CsACTIN1 | |
| Recommended comprehensive ranking | CsUBC1 | CsPTB1 | CsCLATHRIN1 | CsEF1 | CsSAND1 | CsTIP1 | CsACTIN1 | CsGAPDH1 | Cs18S rRNA1 | CsUBQ1 | CsACTIN1 |
| All Samples Ranking Order | |||||||||||
| ∆CT | CsEF1 | CsCLATHRIN1 | CsGAPDH1 | CsPTB1 | CsUBC1 | CsSAND1 | CsTIP1 | CsUBQ1 | Cs18S rRNA1 | CsACTIN1 | CsACTIN1 |
| BestKeeper | CsPTB1 | CsSAND1 | CsUBC1 | CsTIP1 | CsEF1 | CsGAPDH1 | CsCLATHRIN1 | Cs18S rRNA1 | CsUBQ1 | CsACTIN1 | CsACTIN1 |
| Normfinder | CsCLATHRIN1 | CsEF1 | CsGAPDH1 | CsUBC1 | CsPTB1 | CsUBQ1 | CsTIP1 | CsSAND1 | Cs18S rRNA1 | CsACTIN1 | CsACTIN1 |
| Genorm | CsPTB1|CsSAND1 | CsUBC1 | CsTIP1 | CsEF1 | CsGAPDH1 | CsCLATHRIN1 | CsUBQ1 | Cs18S rRNA1 | CsACTIN1 | CsACTIN1 | |
| Recommended comprehensive ranking | CsPTB1 | CsEF1 | CsSAND1 | CsCLATHRIN1 | CsUBC1 | CsGAPDH1 | CsTIP1 | CsUBQ1 | Cs18S rRNA1 | CsACTIN1 | CsACTIN1 |
4. Experimental Section
4.1. Plant Materials and Treatments
4.2. Total RNA Extraction and cDNA Template Preparation
4.3. Primer Design and Quantitative Real-Time PCR (qRT-PCR)
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bustin, S.A.; Benes, V.; Nolan, T.; Pfaffl, M.W. Quantitative real-time RT-PCR—A perspective. J. Mol. Endocrinol. 2005, 34, 597–601. [Google Scholar] [CrossRef] [PubMed]
- Ginzinger, D.G. Gene quantification using real-time quantitative PCR: An emerging technology hits the mainstream. Exp. Hematol. 2002, 30, 503–512. [Google Scholar] [CrossRef] [PubMed]
- Klein, D. Quantification using real-time PCR technology: Applications and limitations. Trends Mol. Med. 2002, 8, 257–260. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A-Z of Quantitative PCR; International University Line: La Jolla, CA, USA, 2004; pp. 87–112. [Google Scholar]
- Nolan, T.; Hands, R.E.; Bustin, S.A. Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 2006, 1, 1559–1582. [Google Scholar] [CrossRef] [PubMed]
- Wong, M.L.; Medrano, J.F. Real-time PCR for mRNA quantitation. Biotechniques 2005, 39, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Huggett, J.; Dheda, K.; Bustin, S.; Zumla, A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005, 6, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Chao, W.S.; Doğramaci, M.; Foley, M.E.; Horvath, D.P.; Anderson, J.V. Selection and validation of endogenous reference genes for qRT-PCR analysis in leafy spurge (Euphorbia esula). PLoS One 2012, 7, e42839. [Google Scholar] [CrossRef] [PubMed]
- Thellin, O.; Zorzi, W.; Lakaye, B.; de Borman, B.; Coumans, B.; Hennen, G.; Grisar, T.; Igout, A.; Heinen, E. Housekeeping genes as internal standards: Use and limits. J. Biotechnol. 1999, 75, 291–295. [Google Scholar] [CrossRef] [PubMed]
- Graham, H.N. Green tea composition, consumption, and polyphenol chemistry. Prev. Med. 1992, 21, 334–350. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.S.; Landau, J.M. Effects of tea consumption on nutrition and health. J. Nutr. 2000, 130, 2409–2412. [Google Scholar] [PubMed]
- Fujiki, H.; Suganuma, M.; Imai, K.; Nakachi, K. Green tea: Cancer preventive beverage and/or drug. Cancer Lett. 2002, 188, 9–13. [Google Scholar] [CrossRef] [PubMed]
- Basu, A.; Sanchez, K.; Leyva, M.J.; Wu, M.; Betts, N.M.; Aston, C.E.; Lyons, T.J. Green tea supplementation affects body weight, lipids, and lipid peroxidation in obese subjects with metabolic syndrome. J. Am. Coll. Nutr. 2010, 29, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Mondal, T.K.; Bhattacharya, A.; Laxmikumaran, M.; Ahuja, P.S. Recent advances of tea (Camellia Sinensis) biotechnology. Plant Cell Tissue Organ Cult. 2004, 76, 195–254. [Google Scholar] [CrossRef]
- Singh, K.; Raizada, J.; Bhardwaj, P.; Ghawana, S.; Rani, A.; Singh, H.; Kaul, K.; Kumar, S. 26s rRNA-based internal control gene primer pair for reverse transcription-polymerase chain reaction-based quantitative expression studies in diverse plant species. Anal. Biochem. 2004, 335, 330–333. [Google Scholar] [CrossRef] [PubMed]
- Gohain, B.; Bandyopadhyay, T.; Borchetia, S.; Bharalee, R.; Gupta, S.; Bhorali, P.; Agarwala, N.; Das, S. Identification and validation of stable reference genes in camellia species. J. Biotechnol. Pharm. Res. 2011, 2, 9–18. [Google Scholar]
- Wang, Y.; Jiang, C.-J.; Li, Y.-Y.; Wei, C.-L.; Deng, W.-W. CsICE1 and csCBF1: Two transcription factors involved in cold responses in Camellia sinensis. Plant Cell. Rep. 2012, 31, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.; Kumar, S.; Ahuja, P.S. Differential expression of histone H3 gene in tea (Camellia sinensis (L.) O. Kuntze) suggests its role in growing tissue. Mol. Biol. Rep. 2009, 36, 537–542. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-C.; Zhao, Q.-Y.; Ma, C.-L.; Zhang, Z.-H.; Cao, H.-L.; Kong, Y.-M.; Yue, C.; Hao, X.-Y.; Chen, L.; Ma, J.-Q. Global transcriptome profiles of Camellia sinensis during cold acclimation. BMC Genomics 2013, 14. [Google Scholar] [CrossRef]
- Das, A.; Das, S.; Mondal, T.K. Identification of differentially expressed gene profiles in young roots of tea [Camellia sinensis (L.) O. Kuntze] subjected to drought stress using suppression subtractive hybridization. Plant Mol. Biol. Rep. 2012, 30, 1088–1101. [Google Scholar] [CrossRef]
- Wang, X.; Hao, X.; Ma, C.; Cao, H.; Yue, C.; Wang, L.; Zeng, J.; Yang, Y. Identification of differential gene expression profiles between winter dormant and sprouting axillary buds in tea plant (Camellia sinensis) by suppression subtractive hybridization. Tree Genet. Genomes 2014, 10, 1149–1159. [Google Scholar] [CrossRef]
- Wang, L.; Li, X.; Zhao, Q.; Jing, S.; Chen, S.; Yuan, H. Identification of genes induced in response to low-temperature treatment in tea leaves. Plant Mol. Biol. Rep. 2009, 27, 257–265. [Google Scholar] [CrossRef]
- Paul, A.; Kumar, S. Responses to winter dormancy, temperature, and plant hormones share gene networks. Funct. Integr. Genomics 2011, 11, 659–664. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.-Y.; Yang, H.; Wei, C.-L.; Yu, O.; Zhang, Z.-Z.; Jiang, C.-J.; Sun, J.; Li, Y.-Y.; Chen, Q.; Xia, T. Deep sequencing of the Camellia sinensis transcriptome revealed candidate genes for major metabolic pathways of tea-specific compounds. BMC Genomics 2011, 12. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 2000, 25, 169–193. [Google Scholar] [CrossRef] [PubMed]
- Dheda, K.; Huggett, J.F.; Bustin, S.A.; Johnson, M.A.; Rook, G.; Zumla, A. Validation of housekeeping genes for normalizing rna expression in real-time PCR. Biotechniques 2004, 37, 112–119. [Google Scholar] [PubMed]
- Czechowski, T.; Stitt, M.; Altmann, T.; Udvardi, M.K.; Scheible, W.-R. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005, 139, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Ailenberg, M.; Silverman, M. Semiquantitative RT-PCR: Enhancement of assay accuracy and reproducibility. Biotechniques 1997, 22, 630–636. [Google Scholar] [PubMed]
- Lee, J.H.; Yoo, S.J.; Park, S.H.; Hwang, I.; Lee, J.S.; Ahn, J.H. Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev. 2007, 21, 397–402. [Google Scholar] [CrossRef] [PubMed]
- Pin, P.; Nilsson, O. The multifaceted roles of FLOWERING LOCUS T in plant development. Plant Cell Environ. 2012, 35, 1742–1755. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Okamoto, T. Involvement of polypyrimidine tract-binding protein (ptb)-related proteins in pollen germination in Arabidopsis. Plant Cell Phys. 2009, 50, 179–190. [Google Scholar] [CrossRef]
- Cottage, A.; Edwards, Y.J.; Elgar, G. SAND, a new protein family: From nucleic acid to protein structure and function prediction. Comp. Funct. Genomics 2001, 2, 226–235. [Google Scholar] [CrossRef] [PubMed]
- Poteryaev, D.; Spang, A. A role of SAND-family proteins in endocytosis. Biochem. Soc. Trans. 2005, 33, 606–608. [Google Scholar] [CrossRef] [PubMed]
- Remans, T.; Smeets, K.; Opdenakker, K.; Mathijsen, D.; Vangronsveld, J.; Cuypers, A. Normalisation of real-time RT-PCR gene expression measurements in Arabidopsis thaliana exposed to increased metal concentrations. Planta 2008, 227, 1343–1349. [Google Scholar] [CrossRef] [PubMed]
- Mafra, V.; Kubo, K.S.; Alves-Ferreira, M.; Ribeiro-Alves, M.; Stuart, R.M.; Boava, L.P.; Rodrigues, C.M.; Machado, M.A. Reference genes for accurate transcript normalization in citrus genotypes under different experimental conditions. PLoS One 2012, 7, e31263. [Google Scholar] [CrossRef] [PubMed]
- Reid, K.E.; Olsson, N.; Schlosser, J.; Peng, F.; Lund, S.T. An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol. 2006, 6. [Google Scholar] [CrossRef] [PubMed]
- Migocka, M.; Papierniak, A. Identification of suitable reference genes for studying gene expression in cucumber plants subjected to abiotic stress and growth regulators. Mol. Breed. 2011, 28, 343–357. [Google Scholar] [CrossRef]
- Artico, S.; Nardeli, S.M.; Brilhante, O.; Grossi-de-Sa, M.F.; Alves-Ferreira, M. Identification and evaluation of new reference genes in Gossypium hirsutum for accurate normalization of real-time quantitative RT-PCR data. BMC Plant Biol. 2010, 10. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhong, H.-Y.; Kuang, J.-F.; Li, J.-G.; Lu, W.-J.; Chen, J.-Y. Validation of reference genes for RT-qPCR studies of gene expression in banana fruit under different experimental conditions. Planta 2011, 234, 377–390. [Google Scholar] [CrossRef] [PubMed]
- Demidenko, N.V.; Logacheva, M.D.; Penin, A.A. Selection and validation of reference genes for quantitative real-time PCR in buckwheat (Fagopyrum esculentum) based on transcriptome sequence data. PLoS One 2011, 6, e19434. [Google Scholar] [CrossRef] [PubMed]
- Shiina, N.; Gotoh, Y.; Kubomura, N.; Iwamatsu, A.; Nishida, E. Microtubule severing by elongation factor 1 alpha. Science 1994, 266, 282–285. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Yu, S.; Zhang, F.; Shen, X.; Zhao, X.; Yu, Y.; Zhang, D. Reference gene selection for real-time quantitative polymerase chain reaction of mRNA transcript levels in chinese cabbage (Brassica rapa L. Ssp. Pekinensis.). Plant Mol. Biol. Rep. 2010, 28, 597–604. [Google Scholar] [CrossRef]
- Xu, M.; Zhang, B.; Su, X.; Zhang, S.; Huang, M. Reference gene selection for quantitative real-time polymerase chain reaction in Populus. Anal. Biochem. 2011, 408, 337–339. [Google Scholar] [CrossRef] [PubMed]
- Fu, W.; Xie, W.; Zhang, Z.; Wang, S.; Wu, Q.; Liu, Y.; Zhou, X.; Zhou, X.; Zhang, Y. Exploring valid reference genes for quantitative real-time PCR analysis in Plutella xylostella (lepidoptera: Plutellidae). Int. J. Biol. Sci. 2013, 9, 792–802. [Google Scholar] [CrossRef] [PubMed]
- An, Y.Q.; McDowell, J.M.; Huang, S.; McKinney, E.C.; Chambliss, S.; Meagher, R.B. Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J. 1996, 10, 107–121. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S. Quantification of mrna using real-time reverse transcription PCR (RT-PCR): Trends and problems. J. Mol. Endocrinol. 2002, 29, 23–39. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, S.R.; Oppenheimer, D.G.; Silflow, C.D.; Snustad, D.P. Characterization of the α-tubulin gene family of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 1987, 84, 5833–5837. [Google Scholar] [CrossRef] [PubMed]
- Tong, Z.; Gao, Z.; Wang, F.; Zhou, J.; Zhang, Z. Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Mol. Biol. 2009, 10. [Google Scholar] [CrossRef] [PubMed]
- Nicot, N.; Hausman, J.-F.O.; Hoffmann, L.; Evers, D. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J. Exper. Bot. 2005, 56, 2907–2914. [Google Scholar] [CrossRef]
- Hayashi, K.-I.; Neve, J.; Hirose, M.; Kuboki, A.; Shimada, Y.; Kepinski, S.; Nozaki, H. Rational design of an auxin antagonist of the SCFTIR1 auxin receptor complex. ACS Chem. Biol. 2012, 7, 590–598. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, K.-I.; Tan, X.; Zheng, N.; Hatate, T.; Kimura, Y.; Kepinski, S.; Nozaki, H. Small-molecule agonists and antagonists of F-box protein-substrate interactions in auxin perception and signaling. Proc. Natl. Acad. Sci. USA 2008, 105, 5632–5637. [Google Scholar] [CrossRef] [PubMed]
- Iino, M. Gravitropism and phototropism of maize coleoptiles: Evaluation of the Cholodny-Went theory through effects of auxin application and decapitation. Plant Cell Physiol. 1995, 36, 361–367. [Google Scholar]
- Chang, S.; Puryear, J.; Cairney, J. A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 1993, 11, 113–116. [Google Scholar] [CrossRef]
- Udvardi, M.K.; Czechowski, T.; Scheible, W.-R. Eleven golden rules of quantitative RT-PCR. Plant Cell 2008, 20, 1736–1737. [Google Scholar] [CrossRef] [PubMed]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hao, X.; Horvath, D.P.; Chao, W.S.; Yang, Y.; Wang, X.; Xiao, B. Identification and Evaluation of Reliable Reference Genes for Quantitative Real-Time PCR Analysis in Tea Plant (Camellia sinensis (L.) O. Kuntze). Int. J. Mol. Sci. 2014, 15, 22155-22172. https://doi.org/10.3390/ijms151222155
Hao X, Horvath DP, Chao WS, Yang Y, Wang X, Xiao B. Identification and Evaluation of Reliable Reference Genes for Quantitative Real-Time PCR Analysis in Tea Plant (Camellia sinensis (L.) O. Kuntze). International Journal of Molecular Sciences. 2014; 15(12):22155-22172. https://doi.org/10.3390/ijms151222155
Chicago/Turabian StyleHao, Xinyuan, David P. Horvath, Wun S. Chao, Yajun Yang, Xinchao Wang, and Bin Xiao. 2014. "Identification and Evaluation of Reliable Reference Genes for Quantitative Real-Time PCR Analysis in Tea Plant (Camellia sinensis (L.) O. Kuntze)" International Journal of Molecular Sciences 15, no. 12: 22155-22172. https://doi.org/10.3390/ijms151222155
APA StyleHao, X., Horvath, D. P., Chao, W. S., Yang, Y., Wang, X., & Xiao, B. (2014). Identification and Evaluation of Reliable Reference Genes for Quantitative Real-Time PCR Analysis in Tea Plant (Camellia sinensis (L.) O. Kuntze). International Journal of Molecular Sciences, 15(12), 22155-22172. https://doi.org/10.3390/ijms151222155






