Expression Analysis of Immune Related Genes Identified from the Coelomocytes of Sea Cucumber (Apostichopus japonicus) in Response to LPS Challenge
Abstract
:1. Introduction
2. Results and Discussion
| Gene Name | Transcript ID | 4 h | 24 h | 72 h |
|---|---|---|---|---|
| Pathogen Recognition | ||||
| CD36-like protein | CL21862.Contig1_haishen | −11.13 | 1.47 | −1.44 |
| Cytosol-type hsp70 | CL15292.Contig1_haishen | 1.53 | 20.97 | 3.39 |
| Fucolectin-7-like | CL7223.Contig2_haishen | 13.45 | −8.63 | −4.76 |
| Fibrinogen-like protein | CL220.Contig10_haishen | 2.58 | 1.70 | 2.32 |
| Heat shock protein 90 | Unigene44996_haishen | 2.93 | 4.49 | 6.18 |
| Lipopolysaccharide-binding protein | CL17187.Contig3_haishen | 1.79 | −1.99 | −2.75 |
| Mannan-binding C-type lectin | CL3438.Contig1_haishen | 1.16 | −1.23 | −2.35 |
| Scavenger receptor cysteine-rich protein type 12 precursor | CL14054.Contig1_haishen | 22.16 | 39.40 | 6.02 |
| Toll-like receptor | CL791.Contig3_haishen | −1.21 | 2.97 | 1.37 |
| Toll-like receptor 3 | CL4619.Contig1_haishen | 1.44 | 1.73 | 1.48 |
| Reorganization of Cytoskeleton | ||||
| Actin | Unigene6143_haishen | 1.92 | −1.19 | −1.67 |
| Amassin 2 precursor | Unigene33380_haishen | −12.73 | −2.31 | −5.90 |
| Amassin 4 precursor | Unigene32576_haishen | −5.62 | −2.50 | −3.51 |
| Focal adhesion kinase | CL4773.Contig4_haishen | −1.30 | −6.11 | 1.27 |
| Myosin V | CL15948.Contig2_haishen | −2.04 | −3.73 | −11.79 |
| Inflammation | ||||
| Complement component 3 | CL9805.Contig1_haishen | −1.61 | 1.31 | 1.09 |
| Complement component 3-2 | Unigene40467_haishen | −1.97 | 1.19 | 10.33 |
| Complement factor B | CL3046.Contig1_haishen | 1.18 | 1.14 | 1.23 |
| Complement factor B-2 | CL3046.Contig2_haishen | −1.41 | 1.51 | 1.26 |
| Complement factor H-like | CL339.Contig6_haishen | 1.00 | 1.17 | 1.31 |
| LPS-induced TNF-α factor | Unigene174_haishen | −1.12 | 1.23 | 1.23 |
| Myeloid differentiation primary response gene 88 | Unigene40451_haishen | 1.18 | 1.11 | 1.32 |
| NF-κB transcription factor Rel | CL9343.Contig1_haishen | 1.32 | 1.39 | 1.30 |
| TBK1- like | CL13483.Contig1_haishen | 1.05 | 1.18 | −1.04 |
| TNF receptor-associated factor 3 | CL13373.Contig2_haishen | −1.13 | 1.83 | −1.07 |
| TNF receptor-associated factor 6 | CL11554.Contig2_haishen | 1.49 | 1.56 | 1.12 |
| Thymosin β | CL4869.Contig1_haishen | −5.82 | −3.32 | −1.32 |
| Apoptosis | ||||
| Caspase 6 | CL16102.Contig1_haishen | 2.53 | 6.15 | 19.29 |
| Caspase 8 | CL18389.Contig1_haishen | −1.06 | 1.28 | 1.16 |
| Cathepsin B | Unigene3030_haishen | 8.86 | 7.67 | 9.38 |
| Lysozyme | Unigene8800_haishen | 5.13 | 3.56 | 1.85 |
2.1. Pathogen Recognition
2.2. Reorganization of Cytoskeleton
2.3. Inflammation Reactions
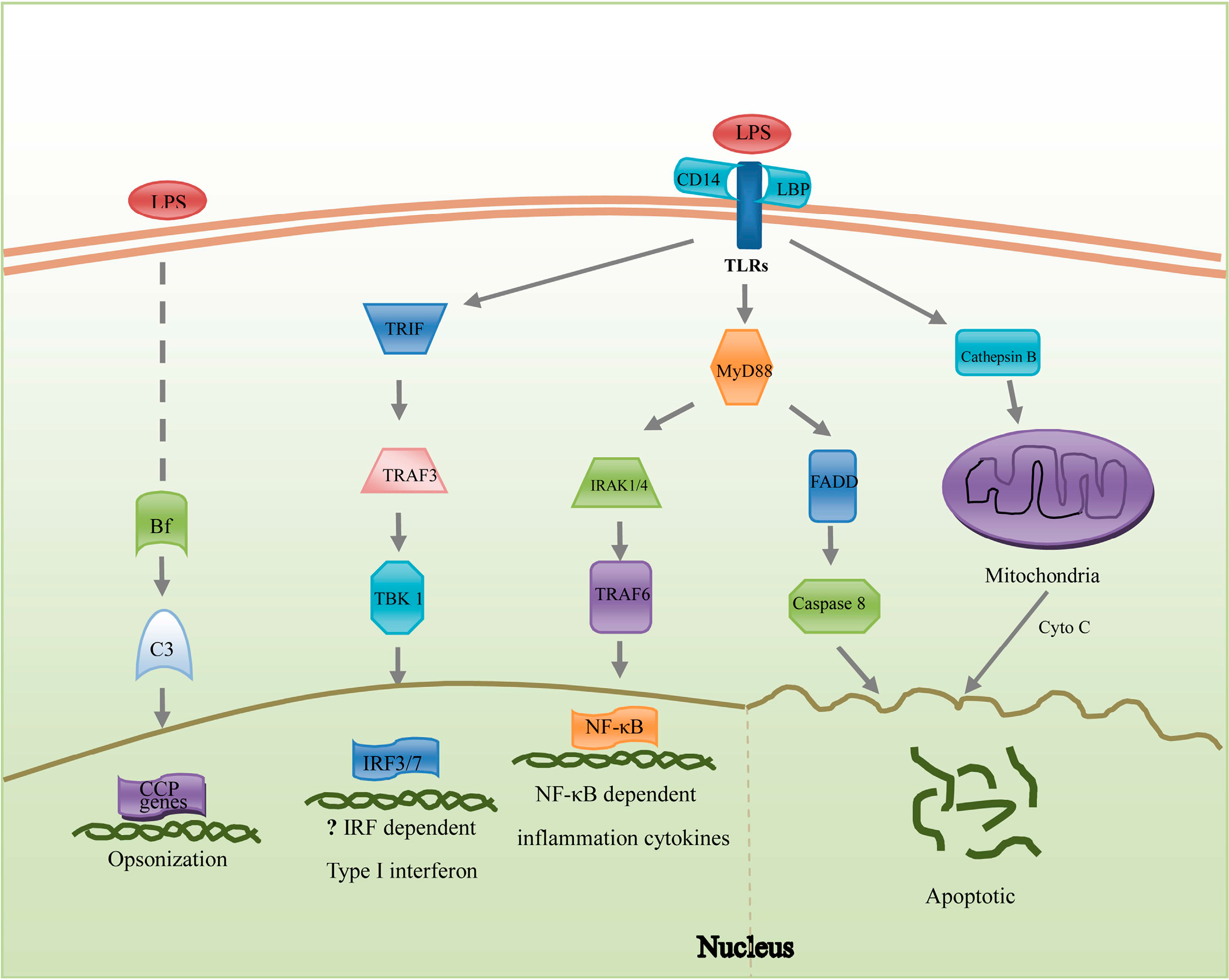
2.4. Apoptosis
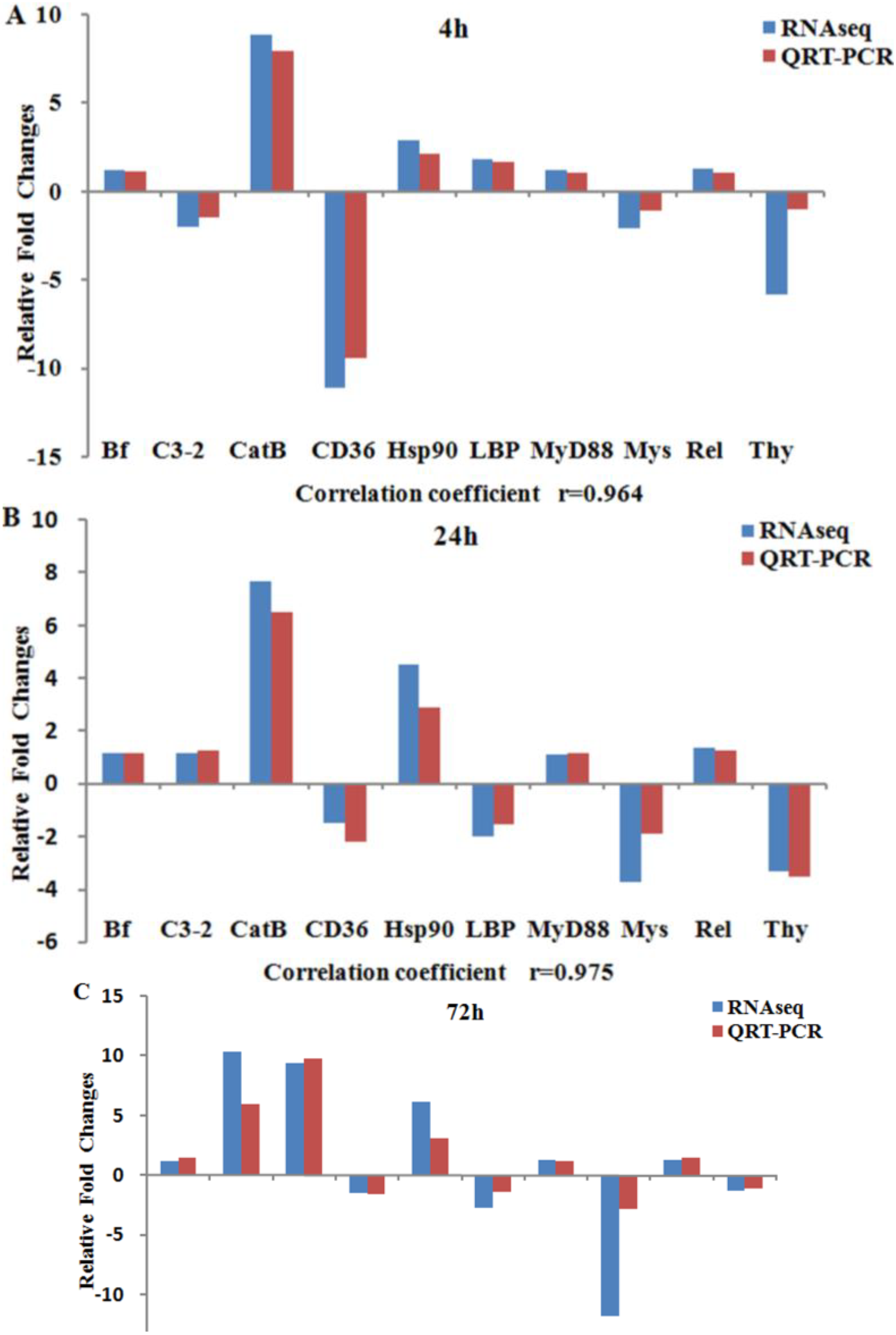
2.5. Validation of Expression Profiles by qRT-PCR
3. Experimental Section
3.1. Sample Collection and RNA Isolation
3.2. cDNA Library Construction and Transcriptome Sequencing
3.3. Analysis of Differentially Expressed Genes
3.4. Expression Validation Using qRT-PCR
| Gene | Primer Sequence (5'-3') |
|---|---|
| Sequencing adaptors | 5-primer: AGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGTA |
| 3-primer: AGATCGGAAGAGCACACGTCTGAACTCCAGTCAC | |
| CD36 | CD36-F: ATTCTTAAAGCCAGCCACA |
| CD36-R: AGTCGTTAGCCGAAGCACC | |
| Complement component 3-2 | C32-F: CTCTCGTGAGTTCTGGC TCAG |
| C32-R: GCAGCCACTGTTACCATCGCGGA | |
| Complement factor B | Bf-F: ATTATCTCGCAACAGCGATCC |
| Bf-R: GGGCAACCACACCGGCTTCTCCA | |
| Cytochrome b | Cytb-F: TGAGCCGCAACAGTAATC |
| Cytb-R: AAGGGAAAAGGAAGTGAAAG | |
| Heat shock protein 90 | Hsp90-F: TATGAAAGCCTGACAGACGCAAGC |
| Hsp90-R: TAACGCAGAGTAAAAGCCAACACC | |
| Lipopolysaccharide-binding protein | LBP-F: AGAAGGGAAATCATACAGAGGCACC |
| LBP-R: TAGCAACATAGTCAGTCATCCACAT | |
| Myosin V | Mys-F: GGGGTGGTCGTCTGATTTGC |
| Mys-R: AAGGTGATTTGAGGAGCGGTA | |
| Myeloid differentiation primary response gene 88 | MyD88-F: CCGATGTAGGAGGATGGTAGTAG |
| MyD88-R: CACAGTAAGGTGCTGAAGAATGC | |
| NF-κB transcription factor Rel | Rel-F: TGCGAAGCCACATCCATT |
| Rel-R: AGGGCATCCTTTAAGTCAGC | |
| Thymosin β | Thy-F: GAGCAGGAGAAAGCAACATAG |
| Thy-R: GAACAAAACAAGCACCCATT |
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hibino, T.; Loza-Coll, M.; Messier, C.; Majeske, A.J.; Cohen, A.H.; Terwilliger, D.P.; Buckley, K.M.; Brockton, V.; Nair, S.V.; Berney, K.; et al. The immune gene repertoire encoded in the purple sea urchin genome. Dev. Biol. 2006, 300, 349–365. [Google Scholar] [CrossRef]
- Rast, J.P.; Smith, L.C.; Loza-Coll, M.; Hibino, T.; Litman, G.W. Genomic insights into the immune system of the sea urchin. Science 2006, 314, 952–956. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Doolittle, R.F. Presence of a vertebrate fibrinogen-like sequence in an echinoderm. Proc. Natl. Acad. Sci. USA 1990, 87, 2097–2101. [Google Scholar] [CrossRef]
- Santiago-Cardona, P.G.; Berrıos, C.A.; Ramırez, F.; Garcıa-Arrarás, J.E. Lipopolysaccharides induce intestinal serum amyloid A expression in the sea cucumber (Holothuria glaberrima). Dev. Comp. Immunol. 2003, 27, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Gómez, F.; Ortíz-Pineda, P.A.; Rojas-Cartagena, C.; Suárez-Castillo, E.C.; García-Ararrás, J.E. Immune-related genes associated with intestinal tissue in the sea cucumber Holothuria glaberrima. Immunogenetics 2008, 60, 57–71. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.L.; Li, C.H.; Wang, D.Q.; Su, X.R.; Jin, C.H.; Li, Y.; Li, T.W. Characterization of two negative regulators of the Toll-like receptor pathway in Apostichopus japonicus: Inhibitor of NF-κB and Toll-interacting protein. Fish Shellfish Immunol. 2013, 35, 1663–1669. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.L.; Li, C.H.; Zhang, P.; Shao, Y.N.; Su, X.R.; Li, Y.; Li, T.W. Two adaptor molecules of MyD88 and TRAF6 in Apostichopus japonicus Toll signaling cascade: Molecular cloning and expression analysis. Dev. Comp. Immunol. 2013, 41, 498–504. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.J.; Zhou, Z.C.; Dong, Y.; Yang, A.F.; Jiang, B.; Gao, S.; Chen, Z.; Guan, X.Y.; Wang, B.; Wang, X.L. Identification and expression analysis of two Toll-like receptor genes from sea cucumber (Apostichopus japonicus). Fish Shellfish Immunol. 2013, 34, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.T.; Sun, Y.X.; Jin, L.J.; Thacker, P.; Li, S.Y.; Xu, Y.P. Aj-rel and Aj-p105, two evolutionary conserved NF-κB homologues in sea cucumber (Apostichopus japonicus) and their involvement in LPS induced immunity. Fish Shellfish Immunol. 2013, 34, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.C.; Sun, D.P.; Yang, A.F.; Dong, Y.; Chen, Z.; Wang, X.Y.; Guan, X.Y.; Jiang, B.; Wang, B. Molecular characterization and expression analysis of a complement component 3 in the sea cucumber (Apostichopus japonicus). Fish Shellfish Immunol. 2011, 31, 540–547. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.; Zhang, F.; Chang, Y.Q. Gene cloning and function analysis of complement B factor-2 of Apostichopus japonicus. Fish Shellfish Immunol. 2012, 33, 504–513. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Yang, H.S.; Zhao, H.L.; Chen, M.Y.; Wang, T.M. The molecular characterization and expression of heat shock protein 90 (Hsp90) and 26 (Hsp26) cDNAs in sea cucumber (Apostichopus japonicus). Cell Stress Chaperones 2011, 16, 481–493. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.Y.; Zhou, Z.C.; Yang, A.F.; Dong, Y.; Chen, Z.; Guan, X.Y.; Jiang, B.; Wang, B. Molecular characterization and expression analysis of heat shock cognate 70 after heat stress and lipopolysaccharide challenge in sea cucumber (Apostichopus japonicus). Biochem. Genet. 2013, 51, 443–457. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.F.; Zhou, Z.C.; Dong, Y.; Jiang, B.; Wang, X.Y.; Chen, Z.; Guan, X.Y.; Wang, B.; Sun, D.P. Expression of immune-related genes in embryos and larvae of sea cucumber Apostichopus japonicus. Fish Shellfish Immunol. 2010, 29, 839–845. [Google Scholar] [CrossRef] [PubMed]
- Bulgakov, A.A.; Eliseikina, M.G.; Petrova, I.Y.; Nazarenko, E.L.; Kovalchuk, S.N.; Kozhemyako, V.B.; Rasskazov, V.A. Molecular and biological characterization of a mannan-binding lectin from the holothurian Apostichopus japonicus. Glycobiology 2007, 17, 1284–1298. [Google Scholar] [CrossRef] [PubMed]
- Fishery Bureau of Ministry of Agriculture PRC. China Fishery Statistical Yearbook, 1st ed.; China Agriculture Press: Beijing, China, 2014; p. 29. [Google Scholar]
- Eeckhaut, I.; Parmentier, E.; Becker, P.; da Silva, S.G.; Jangoux, M. Parasites and biotic diseases in field and cultivated sea cucumbers. Adv. Sea Cucumber Aquac. Manag. 2004, 463, 311–325. [Google Scholar]
- Alexander, C.; Rietschel, E.T. Bacterial lipopolysaccharides and innate immunity. J. Endotoxin Res. 2001, 7, 167–202. [Google Scholar] [PubMed]
- Bannerman, D.D.; Goldblum, S.E. Mechanisms of bacterial lipopolysaccharide-induced endothelial apoptosis. Am. J. Physiol. Lung Cell Mol. Physiol. 2003, 284, L899–L914. [Google Scholar] [PubMed]
- Rowley, A.F.; Powell, A. Invertebrate immune systems-specific, quasispecific, or nonspecific? J. Immunol. 2007, 179, 7209–7214. [Google Scholar] [CrossRef]
- Endean, R. The coelomocytes and coelomic fluids. In Physiol. Echinodermata; Boolootian, R.A., Ed.; Wiley Interscience: New York, NY, USA, 1966; pp. 301–328. [Google Scholar]
- Holm, K.; Dupont, S.; Sköld, H.; Stenius, A.; Thorndyke, M.; Hernroth, B. Induced cell proliferation in putative haematopoietic tissues of the sea star, Asterias rubens. J. Exp. Biol. 2008, 211, 2551–2558. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.C.; Davidson, E.H. The echinoderm immune system. Ann. N. Y. Acad. Sci. 1994, 712, 213–226. [Google Scholar] [CrossRef] [PubMed]
- Johnson, P.T. The coelomic elements of sea urchins (Strongylocentrotus).I. The normal coelomocytes; their morphology and dynamics in hanging drops. J. Invertebr. Pathol. 1969, 13, 42–62. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.C.; Britten, R.J.; Davidson, E.H. SpCoel1: A sea urchin profilin gene expressed specifically in coelomocytes in response to injury. Mol. Biol. Cell 1992, 3, 403–414. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.C.; Davidson, E.H. The echinoid immune system and the phylogenetic occurrence of immune mechanisms in deuterostomes. Immunol. Today 1992, 13, 356–362. [Google Scholar] [CrossRef] [PubMed]
- Edds, K.T. Effects of cytochalasin and colcemid on cortical flow in coelomocytes. Cell Motil. Cytoskelet. 1993, 26, 262–273. [Google Scholar] [CrossRef]
- Gross, P.S.; Al-Sharif, W.Z.; Clow, L.A.; Smith, L.C. Echinoderm immunity and the evolution of the complement system. Dev. Comp. Immunol. 1999, 23, 429–442. [Google Scholar] [CrossRef] [PubMed]
- Gross, P.S.; Clow, L.A.; Smith, L.C. SpC3, the complement homologue from the purple sea urchin, Strongylocentrotus purpuratus, is expressed in two subpopulations of the phagocytic coelomocytes. Immunogenetics 2000, 51, 1034–1044. [Google Scholar] [CrossRef] [PubMed]
- Strickler, S.R.; Bombarely, A.; Mueller, L.A. Designing a transcriptome next generation sequencing project for a nonmodel plant species. Am. J. Bot. 2012, 99, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Ozsolak, F.; Milos, P.M. RNA sequencing: Advances, challenges and opportunities. Nat. Rev. Genet. 2011, 12, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Sadamoto, H.; Takahashi, H.; Okada, T.; Kenmoku, H.; Toyota, M.; Asakawa, Y. De novo sequencing and transcriptome analysis of the centralnervous system of mollusc Lymnaea stagnalis by deep RNA sequencing. PLoS One 2012, 7, e42546. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.N.; Chen, M.Y.; Yang, H.S.; Wang, T.M.; Liu, B.Z.; Shu, C.; Gardiner, D.M. Large scale gene expression profiling during intestine and body wall regeneration in the sea cucumber (Apostichopus japonicus). Comp. Biochem. Physiol. D Genomics Proteomics 2011, 6, 195–205. [Google Scholar] [CrossRef]
- Du, H.X.; Bao, Z.M.; Hou, R.; Wang, S.; Su, H.L.; Yan, J.J.; Tian, M.L.; Li, Y.; Wei, W.; Lu, W.; et al. Transcriptome sequencing and characterization for the sea cucumber Apostichopus japonicus (Selenka, 1867). PLoS One 2012, 7, e33311. [Google Scholar] [CrossRef] [PubMed]
- Li, C.H.; Feng, W.D.; Qiu, L.M.; Xia, C.G.; Su, X.R.; Jin, C.H.; Zhou, T.T.; Zeng, Y.; Li, T.W. Characterization of skin ulceration syndrome associated microRNAs in sea cucumber Apostichopus japonicus by deep sequencing. Fish Shellfish Immunol. 2012, 33, 436–441. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.C.; Dong, Y.; Sun, H.J.; Yang, A.F.; Chen, Z.; Gao, S.; Jiang, J.W.; Guan, X.Y.; Jiang, B.; Wang, B. Transcriptome sequencing of sea cucumber (Apostichopus japonicus) and the identification of gene-associated markers. Mol. Ecol. Resour. 2014, 14, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Gómez, F.; Ortíz-Pineda, P.A.; Rivera-Cardona, G.; García-Ararrás, J.E. LPS-induced genes in intestinal tissue of the sea cucumber Holothuria glaberrima. PLoS One 2009, 4, e6178. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, S.; Gordon, S. The role of scavenger receptors in pathogen recognition and innate immunity. Immunobiology 2004, 209, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Pancer, Z. Dynamic expression of multiple scavenger receptor cysteine-rich genes in coelomocytes of the purple sea urchin. Proc. Natl. Acad. Sci. USA 2000, 97, 13156–13161. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.F.; Yuan, S.C.; Guo, L.; Yu, Y.H.; Li, J.; Wu, T.; Liu, T.; Yang, M.Y.; Wu, K.; Liu, H.L.; et al. Genomic analysis of the immune gene repertoire of amphioxus reveals extraordinary innate complexity and diversity. Genome Res. 2008, 18, 1112–1126. [Google Scholar] [CrossRef] [PubMed]
- Greenwalt, D.E.; Watt, K.W.; So, O.Y.; Jiwani, N. PAS IV, an integral membrane protein of mammary epithelial cells, is related to platelet and endothelial cell CD36 (GP IV). Biochemistry 1990, 29, 7054–7059. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Xu, Y.; Li, L.; Wei, S.; Zhang, S.; Liu, Z. Identification, evolution and expression of a CD36 homolog in the basal chordate amphioxus (Branchiostoma japonicum). Fish Shellfish Immunol. 2013, 34, 546–555. [Google Scholar] [CrossRef] [PubMed]
- Habich, C.; Kempe, K.; van der Zee, R.; Rümenapf, R.; Akiyama, H.; Kolb, H.; Burkart, V. Heat shock protein 60: Specific binding of lipopolysaccharide. J. Immunol. 2005, 174, 1298–1305. [Google Scholar] [CrossRef] [PubMed]
- Shukla, H.D.; Pitha, P.M. Role of Hsp90 in systemic lupus erythematosus and its clinical relevance. Autoimmune Dis. 2012, 2012, 728605. [Google Scholar] [PubMed]
- Hillier, B.J.; Vacquier, V.D. Amassion, an olfactomedin protein, mediates the massive intercellular adhension of sea urchin coelomocytes. J. Cell Biol. 2003, 160, 597–604. [Google Scholar] [CrossRef] [PubMed]
- Hänisch, J.; Kölm, R.; Wozniczka, M.; Bumann, D.; Rottner, K.; Stradal, T.E. Activation of a RhoA/Myosin II-dependent but Arp2/3 complex-independent pathway facilitates Salmonella invasion. Cell Host Microbe 2011, 9, 273–285. [Google Scholar] [CrossRef] [PubMed]
- Guiney, D.G.; Lesnick, M. Targeting of the actin cytoskeleton during infection by Salmonella strains. Clin. Immunol. 2005, 114, 248–255. [Google Scholar] [CrossRef] [PubMed]
- Guttman, J.A.; Finlay, B.B. Tight junctions as targets of infectious agents. Biochim. Biophys. Acta 2009, 1788, 832–841. [Google Scholar] [CrossRef] [PubMed]
- Ulevitch, R.J.; Tobias, P.S. Receptor-dependent mechanisms of cell stimulation by bacterial endotoxin. Annu. Rev. Immunol. 1995, 13, 437. [Google Scholar] [CrossRef] [PubMed]
- Decker, T.; Müller, M.; Stockinger, S. The yin and yang of type I interferon activity in bacterial infection. Nat. Rev. Immunol. 2005, 5, 675–687. [Google Scholar] [CrossRef] [PubMed]
- Fujita, T. Evolution of the lectin-complement pathway and its role in innate immunity. Nat. Rev. Immunol. 2002, 2, 346–353. [Google Scholar] [CrossRef] [PubMed]
- Volanakis, J.E. Participation of C3 and its ligands in complement activation. Curr. Top. Microbiol. Immunol. 1989, 153, 1. [Google Scholar]
- Eliseikina, M.; Timchenko, N.; Bulgakov, A.; Magarlamov, T.; Petrova, I. Influence of Yersinia pseudotuberculosis on the immunity of echinoderms. Adv. Exp. Med. Biol. 2003, 529, 173–175. [Google Scholar] [PubMed]
- Takahashi, A.; Alnemri, E.S.; Lazebnik, Y.A.; Fernandes-Alnemri, T.; Litwack, G.; Moir, R.D.; Goldman, R.D.; Poirier, G.G.; Kaufmann, S.H.; Earnshaw, W.C. Cleavage of lamin A by Mch2 α but not CPP32: Multiple interleukin 1 β-converting enzyme-related proteases with distinct substrate recognition properties are active in apoptosis. Proc. Natl. Acad. Sci. USA 1996, 93, 8395–8400. [Google Scholar] [CrossRef] [PubMed]
- Graham, R.K.; Ehrnhoefer, D.E.; Hayden, M.R. Caspase-6 and neurodegeneration. Trends Neurosci. 2011, 34, 646–656. [Google Scholar] [CrossRef] [PubMed]
- Kingham, P.J.; Pocock, J.M. Microglial secreted cathepsin B induces neuronal apoptosis. J. Neurochem. 2001, 76, 1475–1484. [Google Scholar] [CrossRef] [PubMed]
- Guicciardi, M.E.; Deussing, J.; Miyoshi, H.; Bronk, S.F.; Svingen, P.A.; Peters, C.; Kaufmann, S.H.; Gores, G.J. Cathepsin B contributes to TNF-α-mediated hepatocyte apoptosis by promoting mitochondrial release of cytochrome c. J. Clin. Investig. 2000, 106, 1127–1137. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhang, Y.; Wang, R.J.; Lu, J.G.; Nandi, S.; Mohanty, S.; Terhune, J.; Liu, Z.J.; Peatman, E. RNA-Seq analysis of mucosal immune responses reveals signatures of intestinal barrier disruption and pathogen entry following Edwardsiella ictaluri infection in channel catfish, Ictalurus punctatus. Fish Shellfish Immunol. 2012, 32, 816–827. [Google Scholar] [CrossRef] [PubMed]
- Aparicio, G.; Gotz, S.; Conesa, A.; Segrelles, D.; Blanquer, I.; García, J.M.; Hernandez, V.; Robles, M.; Talon, M. Blast2GO goes grid: Developing a grid-enabled prototype for functional genomics analysis. Stud. Health Technol. Inform. 2006, 120, 194. [Google Scholar] [PubMed]
- Audic, S.; Claverie, J.M. The significance of digital gene expression profiles. Genome Res. 1997, 7, 986–995. [Google Scholar] [PubMed]
- Benjamini, Y.; Drai, D.; Elmer, G.; Kafkafi, N.; Golani, I. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 2001, 125, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W.; Horgan, G.W.; Dempfle, L. Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002, 30, e36. [Google Scholar] [CrossRef] [PubMed]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, Y.; Sun, H.; Zhou, Z.; Yang, A.; Chen, Z.; Guan, X.; Gao, S.; Wang, B.; Jiang, B.; Jiang, J. Expression Analysis of Immune Related Genes Identified from the Coelomocytes of Sea Cucumber (Apostichopus japonicus) in Response to LPS Challenge. Int. J. Mol. Sci. 2014, 15, 19472-19486. https://doi.org/10.3390/ijms151119472
Dong Y, Sun H, Zhou Z, Yang A, Chen Z, Guan X, Gao S, Wang B, Jiang B, Jiang J. Expression Analysis of Immune Related Genes Identified from the Coelomocytes of Sea Cucumber (Apostichopus japonicus) in Response to LPS Challenge. International Journal of Molecular Sciences. 2014; 15(11):19472-19486. https://doi.org/10.3390/ijms151119472
Chicago/Turabian StyleDong, Ying, Hongjuan Sun, Zunchun Zhou, Aifu Yang, Zhong Chen, Xiaoyan Guan, Shan Gao, Bai Wang, Bei Jiang, and Jingwei Jiang. 2014. "Expression Analysis of Immune Related Genes Identified from the Coelomocytes of Sea Cucumber (Apostichopus japonicus) in Response to LPS Challenge" International Journal of Molecular Sciences 15, no. 11: 19472-19486. https://doi.org/10.3390/ijms151119472




