ToxDBScan: Large-Scale Similarity Screening of Toxicological Databases for Drug Candidates
Abstract
:1. Introduction
2. Results and Discussion
2.1. Gene Fingerprint Extraction
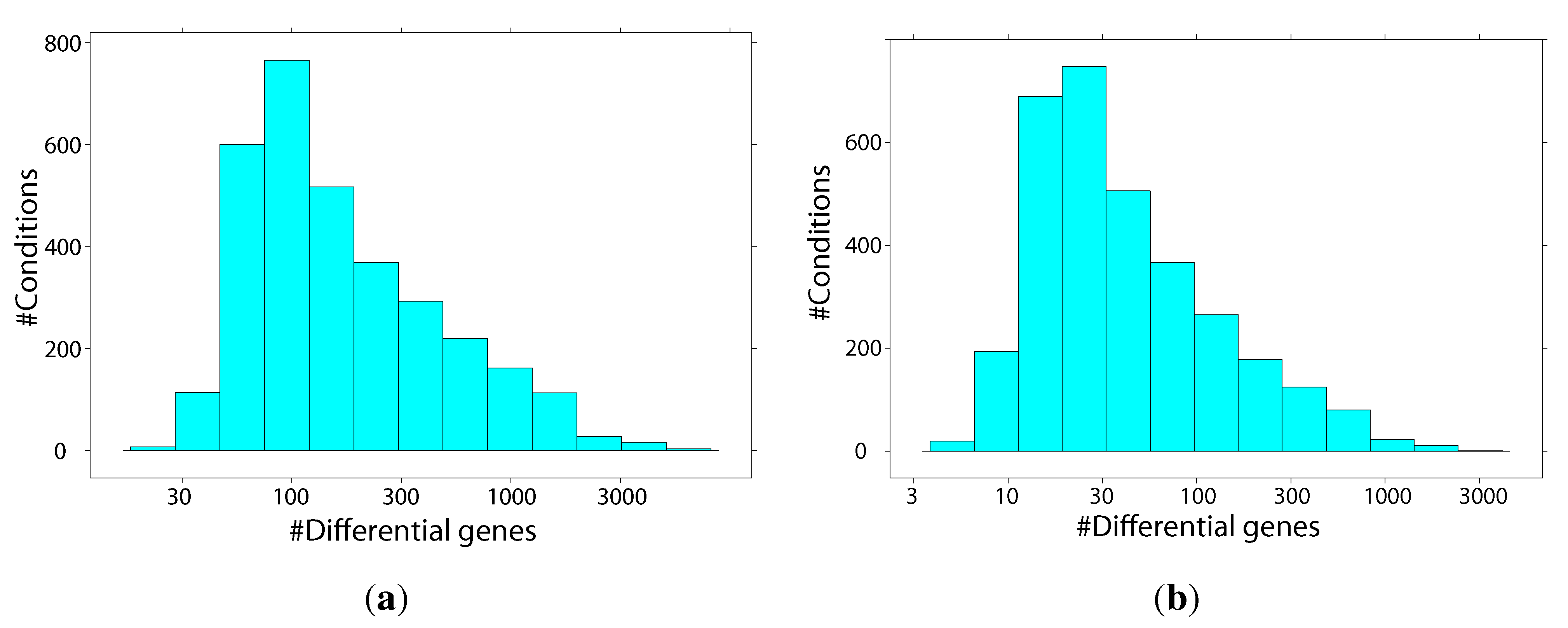
2.2. Identification of Similar Conditions
| Compound | Short Name | CAS Number | Dosing Time (day) | Dose (mg/kg/day) | Contained in |
|---|---|---|---|---|---|
| Genotoxic carcinogens (GCs) | |||||
| Direct Black 38 | CIDB | 1937-37-7 | 7 | 146 | - |
| Nitrosodimethylamine | DMN | 62-75-9 | 7 | 4 | DM |
| Non-genotoxic carcinogens (NGCs) | |||||
| Piperonyl butoxide | PBO | 51-03-6 | 3 | 1200 | - |
| Methyl carbamate | MCA | 598-55-0 | 14 | 400 | - |
| Dehydroepiandrosterone | DHEA | 53-43-0 | 14 | 600 | - |
| Methapyrilene | MP | 135-23-9 | 14 | 60 | TGG, DM |
| Thioacetamide | TAA | 62-55-5 | 7 | 19.2 | TGG, DM |
| Diethylstilbestrol | DES | 56-53-1 | 3 | 10 | DM |
| Wy-14643 | WY | 50892-23-4 | 3 | 60 | TGG, DM |
| Acetamide | AAA | 60-35-5 | 14 | 3000 | TGG |
| Ethionine | ET | 67-21-0 | 14 | 200 | TGG |
| Cyproterone acetate | CPR | 427-51-0 | 14 | 100 | DM |
| Phenobarbital | PB | 50-06-6 | 14 | 80 | TGG, DM |
| Non-hepatocarcinogens (NCs) | |||||
| Cefuroxime | CFX | 55268-75-2 | 14 | 250 | - |
| Nifedipine | NIF | 21829-25-4 | 14 | 3 | TGG |

2.3. Threshold Selection
| Chemical | 1.5-Fold Deregulation | 2-Fold Deregulation | |||||
|---|---|---|---|---|---|---|---|
| Best 5 | Best 10 | Best 20 | Best 5 | Best 10 | Best 20 | ||
| Genotoxic carcinogens | |||||||
| CIDB | 100 | 100 | 95 | 100 | 90 | 90 | |
| DMN | 80 | 70 | 50 | 100 | 80 | 75 | |
| Non-genotoxic carcinogens | |||||||
| PBO | 100 | 80 | 65 | 80 | 70 | 75 | |
| MCA | 60 | 60 | 45 | 80 | 60 | 50 | |
| DHEA | 100 | 90 | 85 | 100 | 90 | 80 | |
| MP | 80 | 70 | 70 | 80 | 50 | 40 | |
| TAA | 100 | 80 | 65 | 100 | 70 | 60 | |
| DES | 100 | 100 | 100 | 100 | 100 | 100 | |
| WY | 100 | 90 | 90 | 100 | 100 | 95 | |
| AAA | 60 | 50 | 35 | 80 | 60 | 50 | |
| ET | 100 | 100 | 100 | 100 | 100 | 85 | |
| CPR | 80 | 80 | 65 | 60 | 60 | 60 | |
| PB | 80 | 60 | 45 | 20 | 20 | 25 | |
| Non-hepatocarcinogens | |||||||
| CFX | 100 | 90 | 85 | 60 | 70 | 85 | |
| NIF | 60 | 80 | 70 | 60 | 70 | 70 | |
| Mean | 86 | 80 | 71 | 82 | 73 | 70 | |
| Chemical | 1.5-Fold Deregulation | 2-Fold Deregulation | |||
|---|---|---|---|---|---|
| S̃ ≥ 0.8 | S̃ ≥ 0.7 | S̃ ≥ 0.8 | S̃ ≥ 0.7 | ||
| Genotoxic carcinogens | |||||
| CIDB | 100 | 100 | 100 | 100 | |
| DMN | 70 | 48 | 78 | 71 | |
| Non-genotoxic carcinogens | |||||
| PBO | 77 | 66 | 73 | 76 | |
| MCA | 100 | 100 | 100 | 100 | |
| DHEA | 88 | 89 | 100 | 80 | |
| MP | 100 | 100 | 100 | 100 | |
| TAA | 100 | 100 | 100 | 67 | |
| DES | 100 | 100 | 100 | 100 | |
| WY | 100 | 91 | 100 | 94 | |
| AAA | 60 | 42 | 67 | 55 | |
| ET | 100 | 100 | 100 | 100 | |
| CPR | 71 | 57 | 50 | 60 | |
| PB | 100 | 60 | 0 | 20 | |
| Non-hepatocarcinogens | |||||
| CFX | 100 | 88 | 33 | 50 | |
| NIF | 50 | 67 | 100 | 100 | |
| Mean | 88 | 80 | 80 | 78 | |
2.4. Hepatocarcinogenicity Prediction
| Chemical | RGC | pGC | RNGC | pNGC | Prediction |
|---|---|---|---|---|---|
| Genotoxic carcinogens | |||||
| CIDB | 1.00 | 0.001 | 0.00 | 1.000 | GC |
| DMN | 0.70 | 0.008 | 0.20 | 0.615 | GC |
| Non-genotoxic carcinogens | |||||
| MP | 0.00 | 1.000 | 1.00 | 0.007 | NGC |
| TAA | 0.00 | 1.000 | 1.00 | 0.012 | NGC |
| DES | 0.00 | 1.000 | 1.00 | <0.001 | NGC |
| WY | 0.00 | 1.000 | 1.00 | <0.001 | NGC |
| PBO | 0.00 | 1.000 | 0.77 | 0.002 | NGC |
| MCA | 0.00 | 1.000 | 1.00 | 0.017 | NGC |
| AAA | 0.00 | 1.000 | 0.60 | 0.048 | NGC |
| DHEA | 0.00 | 1.000 | 0.88 | <0.001 | NGC |
| ET | 0.00 | 1.000 | 1.00 | <0.001 | NGC |
| CPR | 0.00 | 1.000 | 0.71 | 0.018 | NGC |
| PB | 0.00 | 1.000 | 1.00 | 0.019 | NGC |
| Non-hepatocarcinogens | |||||
| CFX | 0.00 | 1.000 | 0.00 | 1.000 | NC |
| NIF | 0.25 | 0.132 | 0.25 | 0.399 | NC |
2.5. Web Application

2.6. Discussion
3. Experimental Section
3.1. Data Resources
3.1.1. Carcinogenic Potency Database
3.1.2. Toxicogenomics Project-Genome Assisted Toxicity Evaluation System
3.1.3. DrugMatrix
3.1.4. Comparison of TG-GATEs and DrugMatrix
3.2. Data Preprocessing
3.3. Pathway Enrichment
3.4. Similarity Scoring
3.5. Performance Evaluation
Conclusions
Supplementary Materials
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Morgan, S.; Grootendorst, P.; Lexchin, J.; Cunningham, C.; Greyson, D. The cost of drug development: A systematic review. Health Policy 2011, 100, 4–17. [Google Scholar] [CrossRef] [PubMed]
- Ames, B.N.; McCann, J.; Yamasaki, E. Methods for detecting carcinogens and mutagens with the salmonella/mammalian-microsome mutagenicity test. Mutat. Res. 1975, 31, 347–363. [Google Scholar] [CrossRef] [PubMed]
- Silva Lima, B.; van der Laan, J.W. Mechanisms of non-genotoxic carcinogenesis and assessment of the human hazard. Regul. Toxicol. Pharmacol. 2000, 32, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Allen, D.G.; Pearse, G.; Haseman, J.K.; Maronpot, R.R. Prediction of rodent carcinogenesis: An evaluation of prechronic liver lesions as forecasters of liver tumors in NTP carcinogenicity studies. Toxicol. Pathol. 2004, 32, 393–401. [Google Scholar] [CrossRef] [PubMed]
- MARCAR—The Project. Available online: http://www.imi-marcar.eu/project.html (accessed on 1 August 2014).
- Liu, Z.; Kelly, R.; Fang, H.; Ding, D.; Tong, W. Comparative analysis of predictive models for non-genotoxic hepatocarcinogenicity using both toxicogenomics and quantitative structure- activity relationships. Chem. Res. Toxicol. 2011, 24, 1062–1070. [Google Scholar] [CrossRef] [PubMed]
- Waters, M.D.; Jackson, M.; Lea, I. Characterizing and predicting carcinogenicity and mode of action using conventional and toxicogenomics methods. Mutat. Res. 2010, 705, 184–200. [Google Scholar] [CrossRef] [PubMed]
- Uehara, T.; Ono, A.; Maruyama, T.; Kato, I.; Yamada, H.; Ohno, Y.; Urushidani, T. The Japanese toxicogenomics project: Application of toxicogenomics. Mol. Nutr. Food Res. 2010, 54, 218–227. [Google Scholar] [CrossRef] [PubMed]
- Ganter, B.; Tugendreich, S.; Pearson, C.I.; Ayanoglu, E.; Baumhueter, S.; Bostian, K.A.; Brady, L.; Browne, L.J.; Calvin, J.T.; Day, G.J.; et al. Development of a large-scale chemogenomics database to improve drug candidate selection and to understand mechanisms of chemical toxicity and action. J. Biotechnol. 2005, 119, 219–244. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. Data, information, knowledge and principle: Back to metabolism in KEGG. Nucleic Acids Res. 2014, 42, D199–D205. [Google Scholar] [CrossRef] [PubMed]
- ZBIT Bioinformatics Toolbox. Available online: http://webservices.cs.uni-tuebingen.de (accessed on 25 August 2014).
- Römer, M.; Eichner, J.; Metzger, U.; Templin, M.F.; Plummer, S.; Ellinger-Ziegelbauer, H.; Zell, A. Cross-platform toxicogenomics for the prediction of non-genotoxic hepatocarcinogenesis in rat. PLoS One 2014, 9, e97640. [Google Scholar] [CrossRef] [PubMed]
- Peraza, M.A.; Burdick, A.D.; Marin, H.E.; Gonzalez, F.J.; Peters, J.M. The toxicology of ligands for peroxisome proliferator-activated receptors (PPAR). Toxicol. Sci. 2006, 90, 269–295. [Google Scholar] [CrossRef] [PubMed]
- Mastrocola, R.; Aragno, M.; Betteto, S.; Brignardello, E.; Catalano, M.G.; Danni, O.; Boccuzzi, G. Pro-oxidant effect of dehydroepiandrosterone in rats is mediated by PPAR activation. Life Sci. 2003, 73, 289–299. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, H.; Shimamoto, K.; Taniai, E.; Ishii, Y.; Morita, R.; Suzuki, K.; Shibutani, M.; Mitsumori, K. Liver tumor promoting effect of omeprazole in rats and its possible mechanism of action. J. Toxicol. Sci. 2012, 37, 491–501. [Google Scholar] [CrossRef] [PubMed]
- Uehara, T.; Minowa, Y.; Morikawa, Y.; Kondo, C.; Maruyama, T.; Kato, I.; Nakatsu, N.; Igarashi, Y.; Ono, A.; Hayashi, H.; et al. Prediction model of potential hepatocarcinogenicity of rat hepatocarcinogens using a large-scale toxicogenomics database. Toxicol. Appl. Pharmacol. 2011, 255, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, J.A.; Hickman, P.; Kimbrough, R.D. Effects of purified and technical piperonyl butoxide on drug-metabolizing enzymes and ultrastructure of rat liver. Toxicol. Appl. Pharmacol. 1973, 26, 444–458. [Google Scholar] [CrossRef] [PubMed]
- Schulte-Hermann, R.; Hoffman, V.; Parzefall, W.; Kallenbach, M.; Gerhardt, A.; Schuppler, J. Adaptive responses of rat liver to the gestagen and anti-androgen cyproterone acetate and other inducers. II. Induction of growth. Chem. Biol. Interact. 1980, 31, 287–300. [Google Scholar] [CrossRef] [PubMed]
- Waxman, D.; Ko, A.; Walsh, C. Regioselectivity and stereoselectivity of androgen hydroxylations catalyzed by cytochrome P-450 isozymes purified from phenobarbital-induced rat liver. J. Biol. Chem. 1983, 258, 11937–11947. [Google Scholar] [PubMed]
- Goecks, J.; Nekrutenko, A.; Taylor, J. Galaxy: A comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010, 11. [Google Scholar] [CrossRef] [PubMed]
- Blankenberg, D.; Von Kuster, G.; Coraor, N.; Ananda, G.; Lazarus, R.; Mangan, M.; Nekrutenko, A.; Taylor, J. Galaxy: A web-based genome analysis tool for experimentalists. Curr. Protocols Mol. Biol. 2010. [Google Scholar] [CrossRef]
- Giardine, B.; Riemer, C.; Hardison, R.C.; Burhans, R.; Elnitski, L.; Shah, P.; Zhang, Y.; Blankenberg, D.; Albert, I.; Taylor, J.; et al. Galaxy: A platform for interactive large-scale genome analysis. Genome Res. 2005, 15, 1451–1455. [Google Scholar] [CrossRef] [PubMed]
- Laulederkind, S.J.F.; Hayman, G.T.; Wang, S.J.; Smith, J.R.; Lowry, T.F.; Nigam, R.; Petri, V.; de Pons, J.; Dwinell, M.R.; Shimoyama, M.; et al. The Rat Genome Database 2013–Data, tools and users. Brief. Bioinf. 2013, 14, 520–526. [Google Scholar] [CrossRef]
- Maglott, D.; Ostell, J.; Pruitt, K.D.; Tatusova, T. Entrez Gene: Gene-centered information at NCBI. Nucleic Acids Res. 2005, 33, D54–D58. [Google Scholar] [CrossRef] [PubMed]
- Flicek, P.; Amode, M.R.; Barrell, D.; Beal, K.; Billis, K.; Brent, S.; Carvalho-Silva, D.; Clapham, P.; Coates, G.; Fitzgerald, S.; et al. Ensembl 2014. Nucleic Acids Res. 2014, 42, D749–D755. [Google Scholar] [CrossRef] [PubMed]
- The UniProt Consortium. Activities at the Universal Protein Resource (UniProt). Nucleic Acids Res. 2014, 42, D191–D198. [Google Scholar]
- DrugMatrix. Available online. 2014. Available online: https://ntp.niehs.nih.gov/drugmatrix/index.html (accessed on 16 June 2014).
- Open TG-GATEs. Available online: http://toxico.nibio.go.jp (accessed on 17 June 2014).
- The Carcinogenic Potency Database (CPDB). Available online: http://toxnet.nlm.nih.gov/cpdb/ (accessed on 30 June 2014).
- Fitzpatrick, R.B. CPDB: Carcinogenic Potency Database. Med. Ref. Serv. Q. 2008, 27, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Takashima, K.; Mizukawa, Y.; Morishita, K.; Okuyama, M.; Kasahara, T.; Toritsuka, N.; Miyagishima, T.; Nagao, T.; Urushidani, T. Effect of the difference in vehicles on gene expression in the rat liver–Analysis of the control data in the Toxicogenomics Project Database. Life Sci. 2006, 78, 2787–2796. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; Suzek, T.O.; Troup, D.B.; Wilhite, S.E.; Ngau, W.C.; Ledoux, P.; Rudnev, D.; Lash, A.E.; Fujibuchi, W.; Edgar, R. NCBI GEO: Mining millions of expression profiles–database and tools. Nucleic Acids Res. 2005, 33, D562–D566. [Google Scholar] [CrossRef] [PubMed]
- IOM (Institute of Medicine). Emerging Safety Science: Workshop Summary; The National Academies Press: Washington, DC, USA, 2008. [Google Scholar]
- Gautier, L.; Cope, L.; Bolstad, B.M.; Irizarry, R.A. Affy–Analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004, 20, 307–315. [Google Scholar] [CrossRef] [PubMed]
- DrugMatrix FTP Server. 2014. Available online: ftp://anonftp.niehs.nih.gov/drugmatrix/ (accessed on 11 April 2014).
- Durinck, S.; Spellman, P.T.; Birney, E.; Huber, W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009, 4, 1184–1191. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. 1995, 57, 289–300. [Google Scholar]
- Rogers, D.; Hahn, M. Extended-connectivity fingerprints. J. Chem. Inf. Model. 2010, 50, 742–754. [Google Scholar] [CrossRef] [PubMed]
- Levandowsky, M.; Winter, D. Distance between sets. Nature 1971, 234, 34–35. [Google Scholar] [CrossRef]
- Cover, T.; Thomas, J. Elements of Information Theory, 2nd ed; Wiley-Interscience: New York City, NY, USA, 2006. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Römer, M.; Backert, L.; Eichner, J.; Zell, A. ToxDBScan: Large-Scale Similarity Screening of Toxicological Databases for Drug Candidates. Int. J. Mol. Sci. 2014, 15, 19037-19055. https://doi.org/10.3390/ijms151019037
Römer M, Backert L, Eichner J, Zell A. ToxDBScan: Large-Scale Similarity Screening of Toxicological Databases for Drug Candidates. International Journal of Molecular Sciences. 2014; 15(10):19037-19055. https://doi.org/10.3390/ijms151019037
Chicago/Turabian StyleRömer, Michael, Linus Backert, Johannes Eichner, and Andreas Zell. 2014. "ToxDBScan: Large-Scale Similarity Screening of Toxicological Databases for Drug Candidates" International Journal of Molecular Sciences 15, no. 10: 19037-19055. https://doi.org/10.3390/ijms151019037
APA StyleRömer, M., Backert, L., Eichner, J., & Zell, A. (2014). ToxDBScan: Large-Scale Similarity Screening of Toxicological Databases for Drug Candidates. International Journal of Molecular Sciences, 15(10), 19037-19055. https://doi.org/10.3390/ijms151019037





