Orthologs of Human Disease Associated Genes and RNAi Analysis of Silencing Insulin Receptor Gene in Bombyx mori
Abstract
:1. Introduction
2. Results and Discussion
2.1. Orthologs of Diseased-Associated Genes in the B. mori
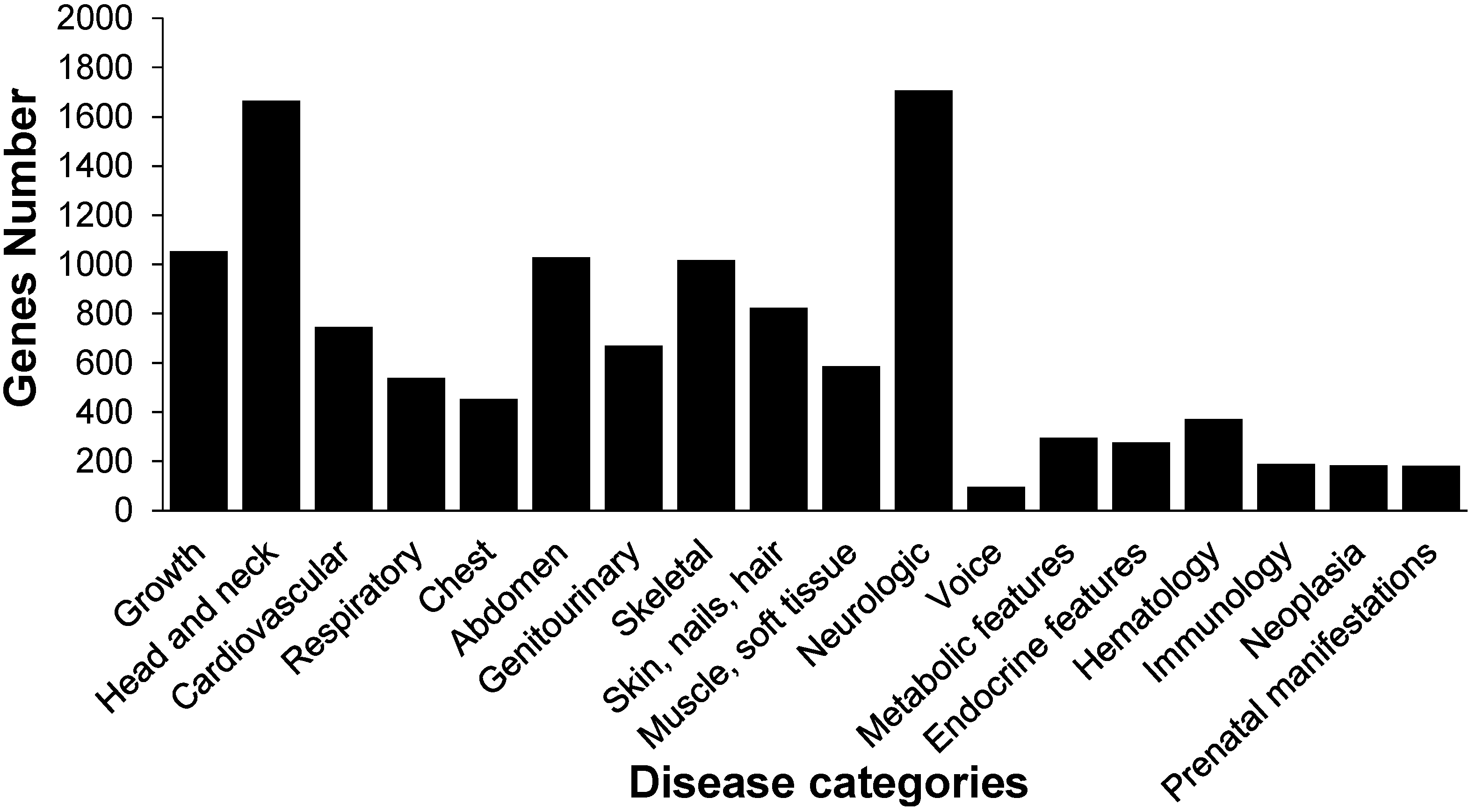

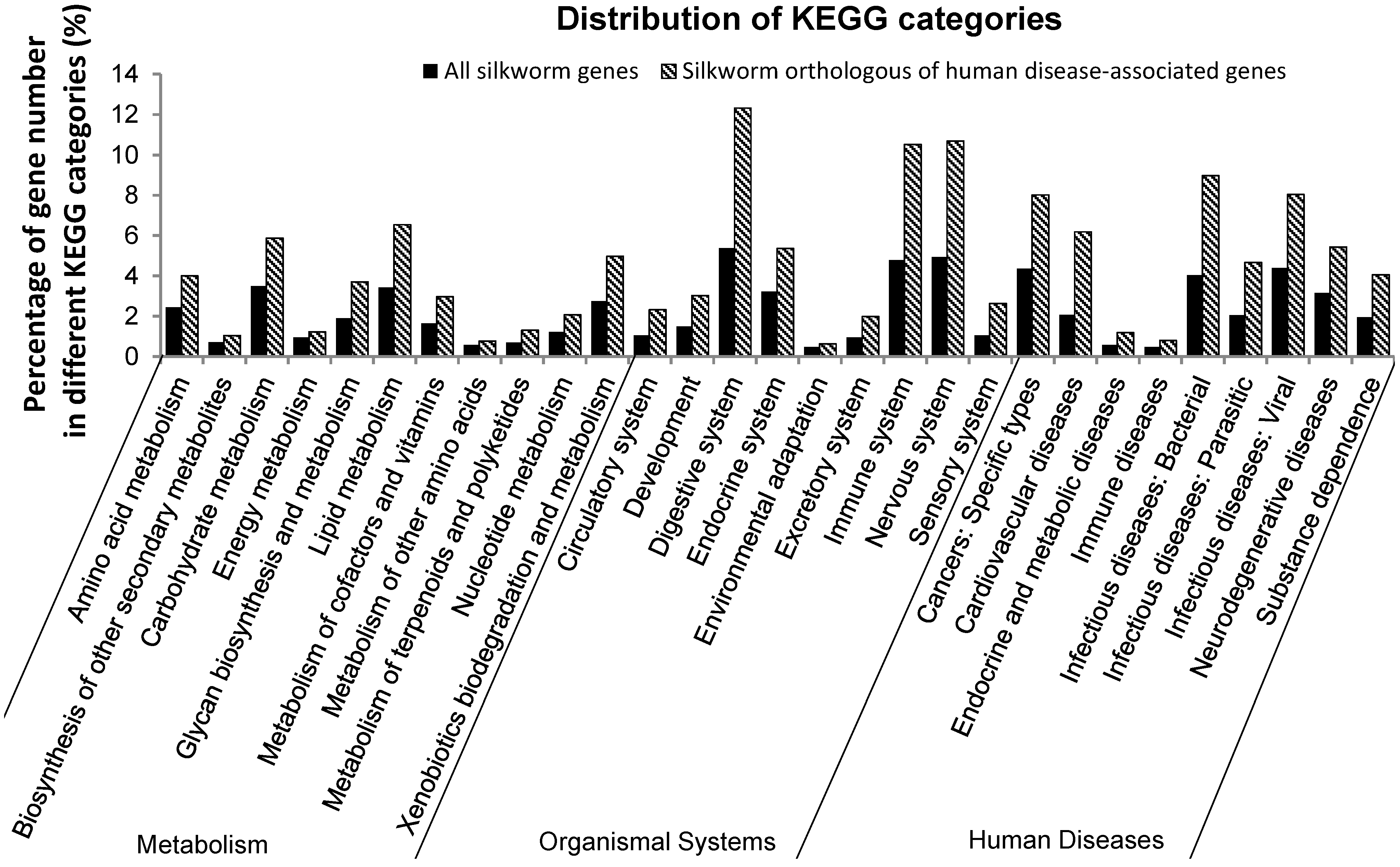
2.2. Insulin Receptor Genes in the B. mori
| Accession Numbers of Human Genes | SilkDB ID | E-Value | Diabetes Disorders |
|---|---|---|---|
| NP_005318.3 | BGIBMGA007360 | 2.00 × 10−89 | Hyperinsulinemic hypoglycemia, familial, 4, 609975. |
| NP_619726.2 | BGIBMGA006839 | 5.00 × 10−28 | Insulin resistance, severe, digenic, 604367. Obesity, resistance to Diabetes, type 2, 125853. |
| EAW69046.1 | BGIBMGA003654 | 0 | Diabetes mellitus, insulin-resistant, with acanthosis nigricans, 610549. Hyperinsulinemic hypoglycemia, familial, 5, 609968. |
| NP_001073285 | BGIBMGA003800 | 2.00 × 10−64 | |
| BAC11220.1 | BGIBMGA009492 | 4.00 × 10−22 | Diabetes mellitus, noninsulin-dependent 1, 601283. |
| NP_000343.2 | BGIBMGA006882 | 9.00 × 10−50 | Diabetes mellitus, noninsulin-dependent, 125853. Diabetes mellitus, permanent neonatal, 606176. Diabetes mellitus, transient neonatal 2, 610374. Hyperinsulinemic hypoglycemia, familial, 1, 256450. |
| NP_005996.2 | BGIBMGA002273 | 1.00 × 10−35 | Diabetes mellitus, noninsulin-dependent, susceptibility to, 125853. |
| BAA91102.1 | BGIBMGA007364 | 1.00 × 10−25 | |
| NP_001007226 | BGIBMGA004315 | 1.00 × 10−8 | |
| NP_006539.3 | BGIBMGA007473 | 1.00 × 10−7 | |
| NP_060244.2 | BGIBMGA011966 | 0 | |
| NP_776250.2 | BGIBMGA005779 | 1.00 × 10−72 | |
| NP_002818.1 | BGIBMGA002096 | 1.00 × 10−66 | |
| AAA52569.1 | BGIBMGA010881 | 1.00 × 10−8 | |
| NP_001033.1 | BGIBMGA002023 | 3.00 × 10−93 | |
| NP_003042.3 | BGIBMGA003138 | 6.00 × 10−40 | Hyperinsulinemic hypoglycemia, familial, 7, 610021. |
| BAF83535.1 | BGIBMGA003740 | 6.00 × 10−44 | Diabetes mellitus, noninsulin-dependent. |
| BAH12783.1 | BGIBMGA004629 | 3.00 × 10−12 | |
| NP_000331.1 | BGIBMGA010740 | 3.00 × 10−51 | |
| NP_005262.1 | BGIBMGA014352 | 1.00 × 10−38 | Hyperinsulinism-hyperammonemia syndrome, 606762. |
| NP_002702.2 | BGIBMGA011660 | 3.00 × 10−19 | Insulin resistance, severe, digenic, 604367. |
| EAX02222.1 | BGIBMGA008597 | 3.00 × 10−22 | Insulin-like growth factor I, resistance to, 270450. |
| NP_000866.1 | BGIBMGA008240 | 3.00 × 10−19 | |
| AAA93480.1 | BGIBMGA003134 | 4.00 × 10−10 | Maturity-onset diabetes of the young 6, 606394. Diabetes mellitus, noninsulin-dependent, 125853. |
| NP_002491.2 | BGIBMGA008390 | 2.00 × 10−9 |
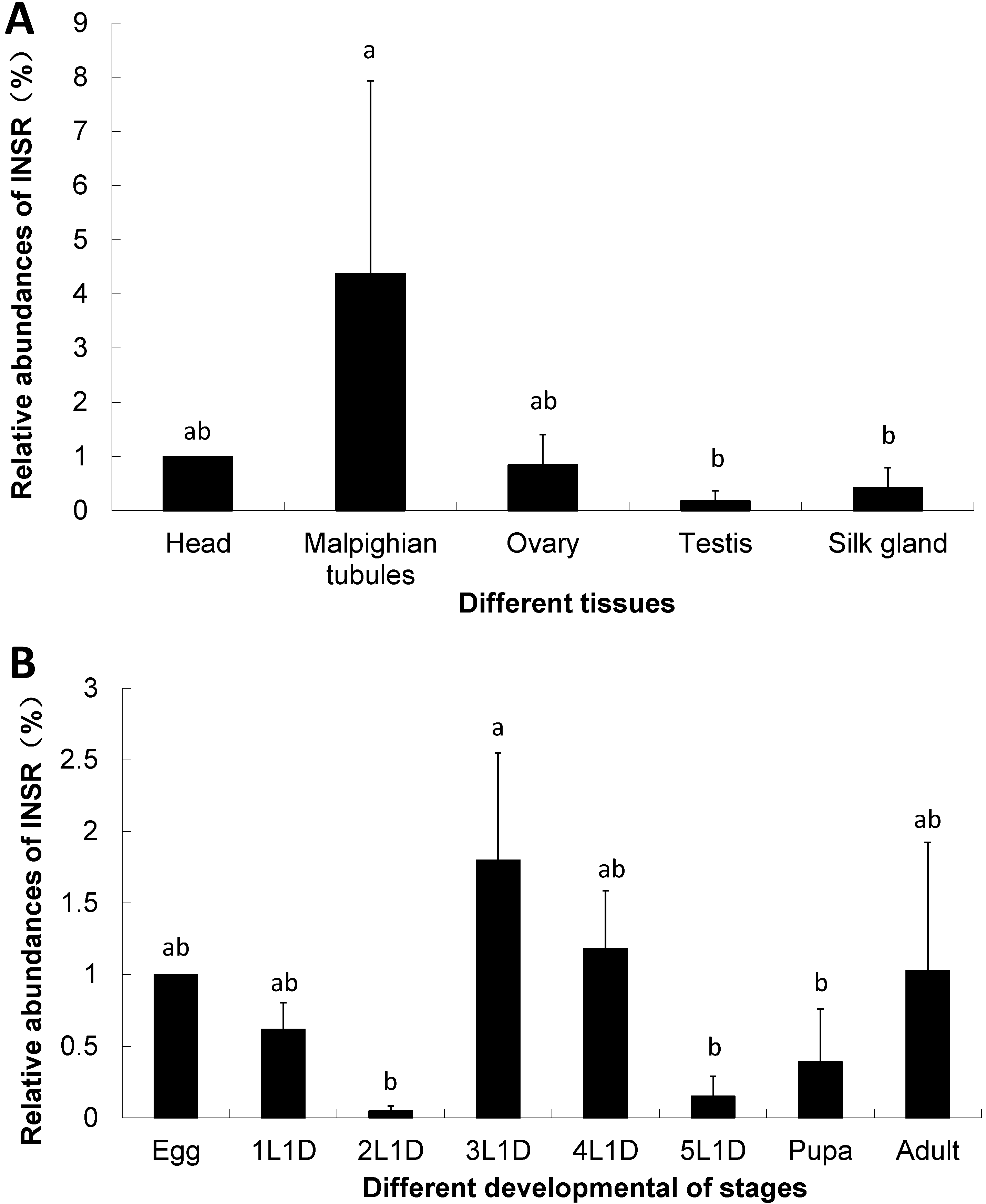
2.3. The Expression of Insulin Receptor Gene in the B. mori
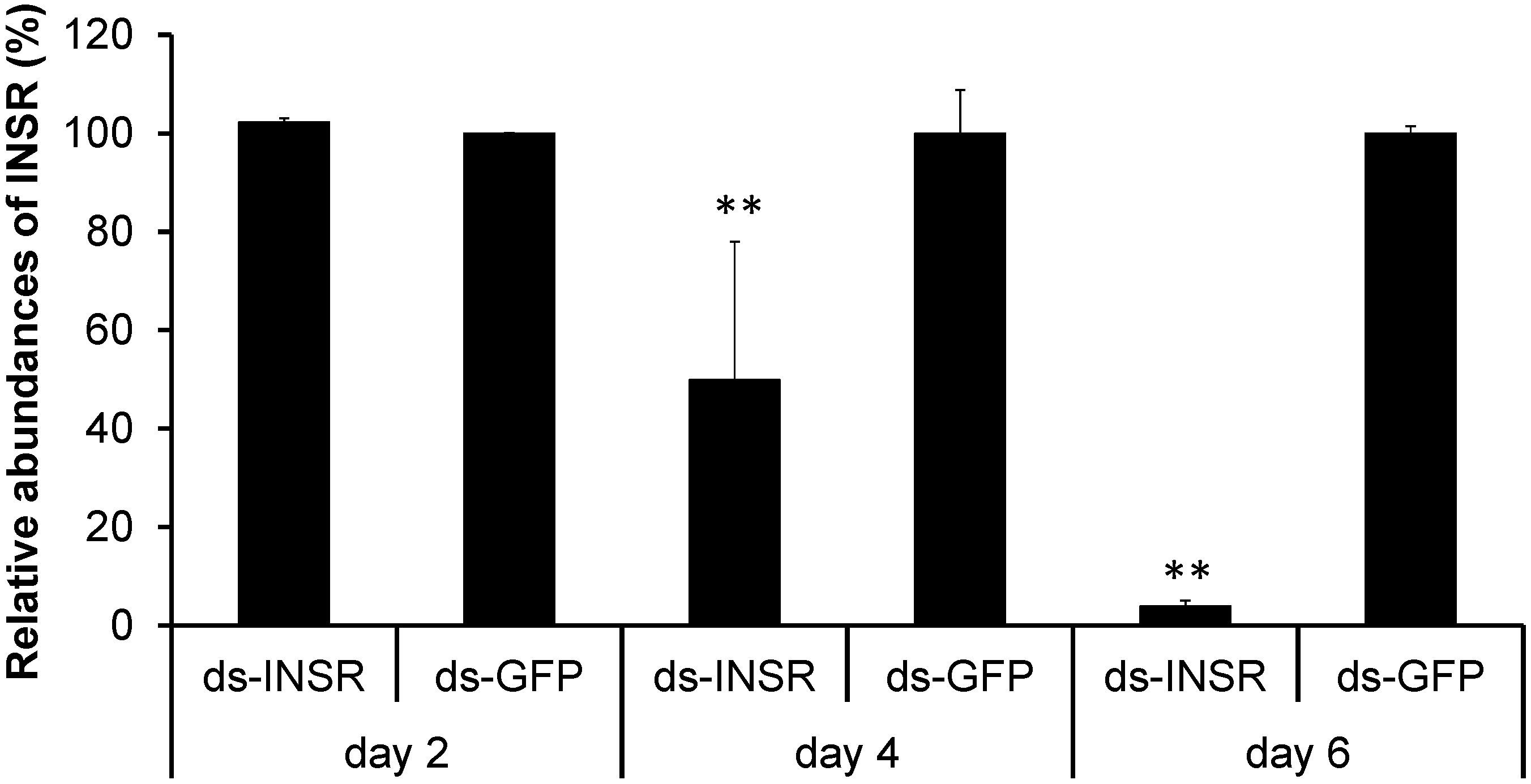
2.4. Knockdown of the Insulin Receptor Genes in the B. mori
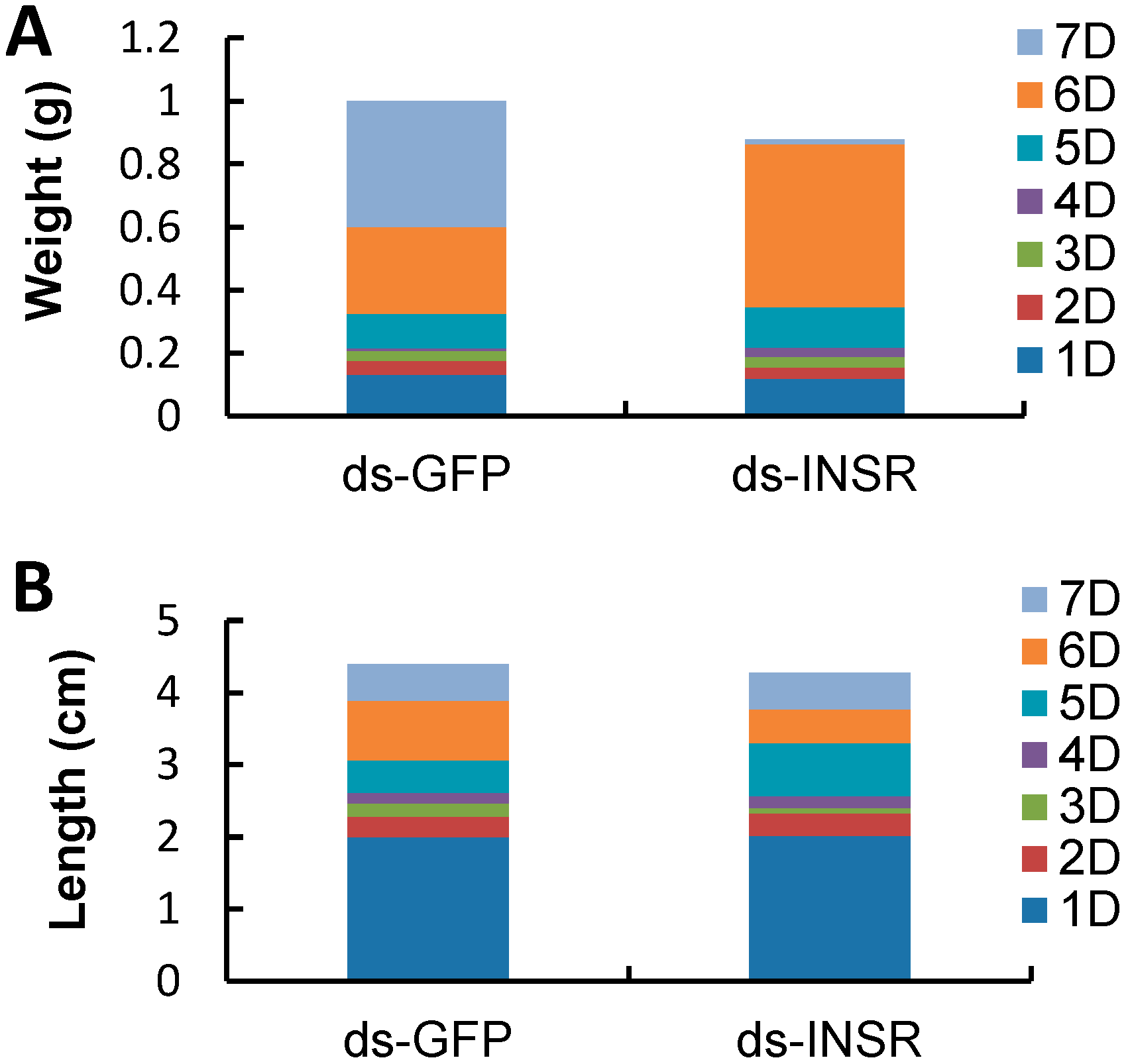
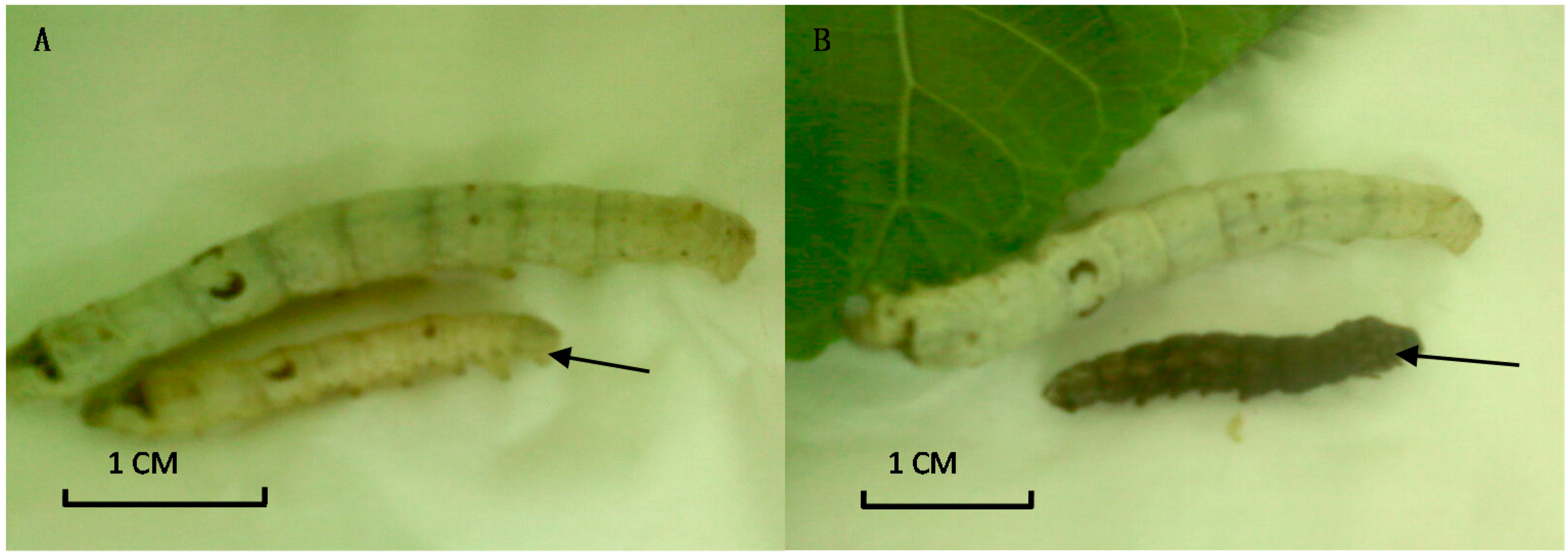
2.5. The Impact of INSR Gene Knockdown on B. mori Development
3. Experimental Section
3.1. Insects
3.2. RNA Purification and cDNA Synthesis
3.3. Quantitative Real-Time PCR
| Genes | Primer Names | Sequences |
|---|---|---|
| Bm-INSR | INSR-qRT-up | 5'-GGTCCGAGATGACTTGTATGG-3' |
| INSR-qRT-down | 5'-GCTGCTGTTGAATGTCTTATAGG-3' | |
| Ribosomal protein 49 | rp49-qRT-up | 5'-AGGCATCAATCGGATCGCTATG-3' |
| rp49-qRT-down | 5'-TTGTGAACTAGGACCTTACGGAATC-3' | |
| Bm-INSR | INSR-dsRNA-up * | 5'-  AGACGCTATCATTGTTGGA-3' AGACGCTATCATTGTTGGA-3' |
| INSR-dsRNA-down | 5'-  TGCGGTTCACTTTACGA-3' TGCGGTTCACTTTACGA-3' | |
| GFP | GFP-dsRNA-up | 5'-  AAGTTCAGCGTGTCCG-3' AAGTTCAGCGTGTCCG-3' |
| GFP-dsRNA-down | 5'-  CACCTTGATGCCGTTC-3' CACCTTGATGCCGTTC-3' |
3.4. Double Strand RNA Synthesis
3.5. DsRNA Injection and Phenotype Observation
3.6. Bioinformatics Analysis
4. Conclusions
Supplementary Materials
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Espinosa-Oliva, A.M.; de Pablos, R.M.; Herrera, A.J. Intracranial injection of lps in rat as animal model of neuroinflammation. Methods Mol. Biol. 2013, 1041, 295–305. [Google Scholar]
- Harrison, C. Animal models: Capturing ad features in a novel rat model. Nat. Rev. Drug Discov. 2013. [Google Scholar] [CrossRef]
- Sasase, T.; Ohta, T.; Masuyama, T.; Yokoi, N.; Kakehashi, A.; Shinohara, M. The spontaneously diabetic torii rat: An animal model of nonobese type 2 diabetes with severe diabetic complications. J. Diabetes Res. 2013, 2013, 976209. [Google Scholar]
- Kreiss, D.S.; Coffman, C.F.; Fiacco, N.R.; Granger, J.C.; Helton, B.M.; Jackson, J.C.; Kim, L.V.; Mistry, R.S.; Mizer, T.M.; Palmer, L.V.; et al. Ritualistic chewing behavior induced by mCPP in the rat is an animal model of obsessive compulsive disorder. Pharmacol. Biochem. Behav. 2013, 104, 119–124. [Google Scholar] [CrossRef]
- Ding, S.; Wu, X.; Li, G.; Han, M.; Zhuang, Y.; Xu, T. Efficient transposition of the piggyBac (Pb) transposon in mammalian cells and mice. Cell 2005, 122, 473–483. [Google Scholar] [CrossRef]
- Marconato, L.; Gelain, M.E.; Comazzi, S. The dog as a possible animal model for human non-Hodgkin lymphoma: A review. Hematol. Oncol. 2013, 31, 1–9. [Google Scholar] [CrossRef]
- Steffan, J.S.; Bodai, L.; Pallos, J.; Poelman, M.; McCampbell, A.; Apostol, B.L.; Kazantsev, A.; Schmidt, E.; Zhu, Y.Z.; Greenwald, M.; et al. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature 2001, 413, 739–743. [Google Scholar] [CrossRef]
- Bilen, J.; Bonini, N.M. Drosophila as a model for human neurodegenerative disease. Annu. Rev. Genet. 2005, 39, 153–171. [Google Scholar] [CrossRef]
- Jung, S.H.; Evans, C.J.; Uemura, C.; Banerjee, U. The Drosophila lymph gland as a developmental model of hematopoiesis. Development 2005, 132, 2521–2533. [Google Scholar] [CrossRef]
- Marsh, J.L.; Pallos, J.; Thompson, L.M. Fly models of Huntington’s disease. Hum. Mol. Genet. 2003, 12, R187–R193. [Google Scholar] [CrossRef]
- Xu, Y.; Condell, M.; Plesken, H.; Edelman-Novemsky, I.; Ma, J.; Ren, M.; Schlame, M. A Drosophila model of barth syndrome. Proc. Natl. Acad. Sci. USA 2006, 103, 11584–11588. [Google Scholar] [CrossRef]
- Whitworth, A.J.; Theodore, D.A.; Greene, J.C.; Benes, H.; Wes, P.D.; Pallanck, L.J. Increased glutathione S-transferase activity rescues dopaminergic neuron loss in a Drosophila model of Parkinson’s disease. Proc. Natl. Acad. Sci. USA 2005, 102, 8024–8029. [Google Scholar] [CrossRef]
- Crowther, D.C.; Kinghorn, K.J.; Miranda, E.; Page, R.; Curry, J.A.; Duthie, F.A.; Gubb, D.C.; Lomas, D.A. Intraneuronal Aβ, non-amyloid aggregates and neurodegeneration in a Drosophila model of Alzheimer’s disease. Neuroscience 2005, 132, 123–135. [Google Scholar] [CrossRef]
- Vidal, M.; Cagan, R.L. Drosophila models for cancer research. Curr. Opin. Genet. Dev. 2006, 16, 10–16. [Google Scholar] [CrossRef]
- Bier, E.; Bodmer, R. Drosophila, an emerging model for cardiac disease. Gene 2004, 342, 1–11. [Google Scholar] [CrossRef]
- Reiter, L.T.; Potocki, L.; Chien, S.; Gribskov, M.; Bier, E. A systematic analysis of human disease-associated gene sequences in Drosophila melanogaster. Genome Res. 2001, 11, 1114–1125. [Google Scholar] [CrossRef]
- Wolf, M.J.; Amrein, H.; Izatt, J.A.; Choma, M.A.; Reedy, M.C.; Rockman, H.A. Drosophila as a model for the identification of genes causing adult human heart disease. Proc. Natl. Acad. Sci. USA 2006, 103, 1394–1399. [Google Scholar] [CrossRef]
- Read, R.D.; Goodfellow, P.J.; Mardis, E.R.; Novak, N.; Armstrong, J.R.; Cagan, R.L. A Drosophila model of multiple endocrine neoplasia type 2. Genetics 2005, 171, 1057–1081. [Google Scholar] [CrossRef]
- Mehta, V.; Peebles, D.; David, A.L. Animal models for prenatal gene therapy: Choosing the right model. Methods Mol. Biol. 2012, 891, 183–200. [Google Scholar]
- Little, C.B.; Zaki, S. What constitutes an “animal model of osteoarthritis”—The need for consensus? Osteoarthr. Cartil. 2012, 20, 261–267. [Google Scholar] [CrossRef]
- Salkovic-Petrisic, M.; Knezovic, A.; Hoyer, S.; Riederer, P. What have we learned from the streptozotocin-induced animal model of sporadic Alzheimer’s disease, about the therapeutic strategies in Alzheimer’s research. J. Neural Transm. 2013, 120, 233–252. [Google Scholar] [CrossRef] [Green Version]
- Xia, Q.; Zhou, Z.; Lu, C.; Cheng, D.; Dai, F.; Li, B.; Zhao, P.; Zha, X.; Cheng, T.; Chai, C.; et al. A draft sequence for the genome of the domesticated silkworm (Bombyx mori). Science 2004, 306, 1937–1940. [Google Scholar] [CrossRef]
- Mita, K.; Kasahara, M.; Sasaki, S.; Nagayasu, Y.; Yamada, T.; Kanamori, H.; Namiki, N.; Kitagawa, M.; Yamashita, H.; Yasukochi, Y.; et al. The genome sequence of silkworm, Bombyx mori. DNA Res. 2004, 11, 27–35. [Google Scholar] [CrossRef]
- Deng, D.; Xu, H.; Wang, F.; Duan, X.; Ma, S.; Xiang, Z.; Xia, Q. The promoter of Bmlp3 gene can direct fat body-specific expression in the transgenic silkworm, Bombyx mori. Transgenic Res. 2013, 22, 1055–1063. [Google Scholar] [CrossRef]
- Jiang, L.; Zhao, P.; Wang, G.; Cheng, T.; Yang, Q.; Jin, S.; Lin, P.; Xiao, Y.; Sun, Q.; Xia, Q. Comparison of factors that may affect the inhibitory efficacy of transgenic RNAi targeting of baculoviral genes in silkworm, Bombyx mori. Antivir. Res. 2013, 97, 255–263. [Google Scholar] [CrossRef]
- Sato, M.; Kojima, K.; Sakuma, C.; Murakami, M.; Aratani, E.; Takenouchi, T.; Tamada, Y.; Kitani, H. Production of scFv-conjugated affinity silk powder by transgenic silkworm technology. PLoS One 2012, 7, e34632. [Google Scholar] [CrossRef]
- Gong, Z.H.; Jin, H.Q.; Jin, Y.F.; Zhang, Y.Z. Expression of cholera toxin B subunit and assembly as functional oligomers in silkworm. J. Biochem. Mol. Biol. 2005, 38, 717–724. [Google Scholar] [CrossRef]
- Du, D.; Kato, T.; Nabi, A.H.; Suzuki, F.; Park, E.Y. Expression of functional human (pro)renin receptor in silkworm (Bombyx mori) larvae using BmMNPV bacmid. Biotechnol. Appl. Biochem. 2008, 49, 195–202. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, Y.; Chen, J.; Zhang, W.; Sheng, Q.; Chen, J.; Yu, W.; Nie, Z.; Zhang, Y.; Wu, W.; et al. A shark liver gene-derived active peptide expressed in the silkworm, Bombyx mori: Preliminary studies for oral administration of the recombinant protein. Mar. Drugs 2013, 11, 1492–1505. [Google Scholar] [CrossRef]
- Tabunoki, H.; Ono, H.; Ode, H.; Ishikawa, K.; Kawana, N.; Banno, Y.; Shimada, T.; Nakamura, Y.; Yamamoto, K.; Satoh, J.; et al. Identification of key uric acid synthesis pathway in a unique mutant silkworm Bombyx mori model of Parkinson’s disease. PLoS One 2013, 8, e69130. [Google Scholar] [CrossRef]
- Meng, Y.; Katsuma, S.; Daimon, T.; Banno, Y.; Uchino, K.; Sezutsu, H.; Tamura, T.; Mita, K.; Shimada, T. The silkworm mutant lemon (lemon lethal) is a potential insect model for human sepiapterin reductase deficiency. J. Biol. Chem. 2009, 284, 11698–11705. [Google Scholar] [CrossRef]
- Matsumoto, Y.; Sumiya, E.; Sugita, T.; Sekimizu, K. An invertebrate hyperglycemic model for the identification of anti-diabetic drugs. PLoS One 2011, 6, e18292. [Google Scholar] [CrossRef]
- Feng, F.; Chen, L.; Lian, C.; Xia, H.; Zhou, Y.; Yao, Q.; Chen, K. Comparative proteomic analysis reveals the suppressive effects of dietary high glucose on the midgut growth of silkworm. J. Proteomics 2014, 108, 124–132. [Google Scholar] [CrossRef]
- Satake, S.; Masumura, M.; Ishizaki, H.; Nagata, K.; Kataoka, H.; Suzuki, A.; Mizoguchi, A. Bombyxin, an insulin-related peptide of insects, reduces the major storage carbohydrates in the silkworm Bombyx mori. Comp. Biochem. Physiol. 1997, 118, 349–357. [Google Scholar] [CrossRef]
- Iwami, M. Bombyxin: An insect brain peptide that belongs to the insulin family. Zool. Sci. 2000, 17, 1035–1044. [Google Scholar] [CrossRef]
- Masumura, M.; Satake, S.; Saegusa, H.; Mizoguchi, A. Glucose stimulates the release of bombyxin, an insulin-related peptide of the silkworm Bombyx mori. Gen. Comp. Endocrinol. 2000, 118, 393–399. [Google Scholar] [CrossRef]
- Online Mendelian Inheritance in Man. Available online: http://omia.angis.org.au/home/ (accessed on 10 November 2011).
- Huang, H.; Winter, E.E.; Wang, H.; Weinstock, K.G.; Xing, H.; Goodstadt, L.; Stenson, P.D.; Cooper, D.N.; Smith, D.; Alba, M.M.; et al. Evolutionary conservation and selection of human disease gene orthologs in the rat and mouse genomes. Genome Biol. 2004, 5, R47. [Google Scholar] [CrossRef] [Green Version]
- Pautsch, A.; Zoephel, A.; Ahorn, H.; Spevak, W.; Hauptmann, R.; Nar, H. Crystal structure of bisphosphorylated IGF-1 receptor kinase: Insight into domain movements upon kinase activation. Structure 2001, 9, 955–965. [Google Scholar] [CrossRef]
- Bansilal, S.; Farkouh, M.E.; Fuster, V. Role of insulin resistance and hyperglycemia in the development of atherosclerosis. Am. J. Cardiol. 2007, 99, 6B–14B. [Google Scholar] [CrossRef]
- Kikuta, S.; Hagiwara-Komoda, Y.; Noda, H.; Kikawada, T. A novel member of the trehalose transporter family functions as an H+-dependent trehalose transporter in the reabsorption of trehalose in malpighian tubules. Front. Physiol. 2012, 3, 290. [Google Scholar] [CrossRef]
- Schertzer, J.D.; Tamrakar, A.K.; Magalhaes, J.G.; Pereira, S.; Bilan, P.J.; Fullerton, M.D.; Liu, Z.; Steinberg, G.R.; Giacca, A.; Philpott, D.J.; et al. Nod1 activators link innate immunity to insulin resistance. Diabetes 2011, 60, 2206–2215. [Google Scholar] [CrossRef]
- Steinberg, G.R. Inflammation in obesity is the common link between defects in fatty acid metabolism and insulin resistance. Cell Cycle 2007, 6, 888–894. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Silkdb. Available online: Ftp://silkdb.org/ (accessed on 12 November 2011).
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Waterhouse, R.M.; Tegenfeldt, F.; Li, J.; Zdobnov, E.M.; Kriventseva, E.V. Orthodb: A hierarchical catalog of animal, fungal and bacterial orthologs. Nucleic Acids Res. 2013, 41, D358–D365. [Google Scholar] [CrossRef]
- Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talon, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef]
- Costa, R.; Speretta, E.; Crowther, D.C.; Cardoso, I. Testing the therapeutic potential of doxycycline in a Drosophila melanogaster model of Alzheimer disease. J. Biol. Chem. 2011, 286, 41647–41655. [Google Scholar] [CrossRef]
- Kim, K.; Kim, S.H.; Kim, J.; Kim, H.; Yim, J. Glutathione S-transferase ω 1 activity is sufficient to suppress neurodegeneration in a Drosophila model of Parkinson disease. J. Biol. Chem. 2012, 287, 6628–6641. [Google Scholar] [CrossRef]
- Qin, H.; Cressy, M.; Li, W.; Coravos, J.S.; Izzi, S.A.; Dubnau, J. γ Neurons mediate dopaminergic input during aversive olfactory memory formation in Drosophila. Curr. Biol. 2012, 22, 608–614. [Google Scholar] [CrossRef]
- Morris, S.N.; Coogan, C.; Chamseddin, K.; Fernandez-Kim, S.O.; Kolli, S.; Keller, J.N.; Bauer, J.H. Development of diet-induced insulin resistance in adult Drosophila melanogaster. Biochim. Biophys. Acta 2012, 1822, 1230–1237. [Google Scholar] [CrossRef]
- Pasco, M.Y.; Leopold, P. High sugar-induced insulin resistance in Drosophila relies on the lipocalin Neural Lazarillo. PLoS One 2012, 7, e36583. [Google Scholar] [CrossRef]
- Warbrick-Smith, J.; Behmer, S.T.; Lee, K.P.; Raubenheimer, D.; Simpson, S.J. Evolving resistance to obesity in an insect. Proc. Natl. Acad. Sci. USA 2006, 103, 14045–14049. [Google Scholar] [CrossRef]
- Dai, L.S.; Zhu, B.J.; Liu, Q.N.; Wei, G.Q.; Liu, C.L. Characterization of the complete mitochondrial genome of Bombyx mori strain h9 (Lepidoptera: Bombycidae). Gene 2013, 519, 326–334. [Google Scholar] [CrossRef]
- Kikuchi, H.; Takahashi, N.; Oshima, Y. Novel aromatics bearing 4-O-methylglucose unit isolated from the oriental crude drug Bombyx Batryticatus. Tetrahedron Lett. 2004, 45, 367–370. [Google Scholar] [CrossRef]
- Hu, K.; Wang, Q.M.; Hu, P.Q. The male silkworm moth (Antheraea pernyi) is a key ingredient in Hu-Bao and Sheng-Bao for specific prolongation of the life-span of the male fruit fly (Drosophila melanogaster). Am. J. Chin. Med. 2002, 30, 263–270. [Google Scholar] [CrossRef]
- Gong, Z.; Jin, Y.; Zhang, Y. Oral administration of a cholera toxin B subunit-insulin fusion protein produced in silkworm protects against autoimmune diabetes. J. Biotechnol. 2005, 119, 93–105. [Google Scholar] [CrossRef]
- Cong, L.; Cao, G.; Renyu, X.; Zhonghua, P.; Xiaojian, Z.; Zhou, W.; Gong, C. Reducing blood glucose level in tidm mice by orally administering the silk glands of transgenic hIGF-I silkworms. Biochem. Biophys. Res. Commun. 2011, 410, 721–725. [Google Scholar] [CrossRef]
- Zhao, H.L.; Tong, P.C.; Chan, J.C. Traditional Chinese medicine in the treatment of diabetes. Nestle Nutr. Workshop Ser. Clin. Perform. Programme 2006, 11, 15–25; discussion 25–19. [Google Scholar]
- Zhang, T.T.; Jiang, J.G. Active ingredients of traditional Chinese medicine in the treatment of diabetes and diabetic complications. Expert Opin. Investig. Drugs 2012, 21, 1625–1642. [Google Scholar] [CrossRef]
- Tong, X.L.; Dong, L.; Chen, L.; Zhen, Z. Treatment of diabetes using traditional Chinese medicine: Past, present and future. Am. J. Chin. Med. 2012, 40, 877–886. [Google Scholar] [CrossRef]
- Kasuli, E.G. Are alternative supplements effective treatment for diabetes mellitus? Nutr. Clin. Pract. 2011, 26, 352–355. [Google Scholar]
- Wu, Q.; Brown, M.R. Signaling and function of insulin-like peptides in insects. Annu. Rev. Entomol. 2006, 51, 1–24. [Google Scholar] [CrossRef]
- Dou, T.; Ji, C.; Gu, S.; Xu, J.; Xu, J.; Ying, K.; Xie, Y.; Mao, Y. Co-evolutionary analysis of insulin/insulin like growth factor 1 signal pathway in vertebrate species. Front. Biosci. 2006, 11, 380–388. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Z.; Teng, X.; Chen, M.; Li, F. Orthologs of Human Disease Associated Genes and RNAi Analysis of Silencing Insulin Receptor Gene in Bombyx mori. Int. J. Mol. Sci. 2014, 15, 18102-18116. https://doi.org/10.3390/ijms151018102
Zhang Z, Teng X, Chen M, Li F. Orthologs of Human Disease Associated Genes and RNAi Analysis of Silencing Insulin Receptor Gene in Bombyx mori. International Journal of Molecular Sciences. 2014; 15(10):18102-18116. https://doi.org/10.3390/ijms151018102
Chicago/Turabian StyleZhang, Zan, Xiaolu Teng, Maohua Chen, and Fei Li. 2014. "Orthologs of Human Disease Associated Genes and RNAi Analysis of Silencing Insulin Receptor Gene in Bombyx mori" International Journal of Molecular Sciences 15, no. 10: 18102-18116. https://doi.org/10.3390/ijms151018102




