Hope for GWAS: Relevant Risk Genes Uncovered from GWAS Statistical Noise
Abstract
:1. Introduction
2. Results
2.1. Functional Connectivity beyond Random Expectation at the Range of Genome-Wide Association Studies (GWAS) Statistical Noise
2.2. Functionally Connected Groups Are Enriched in True Disease Genes
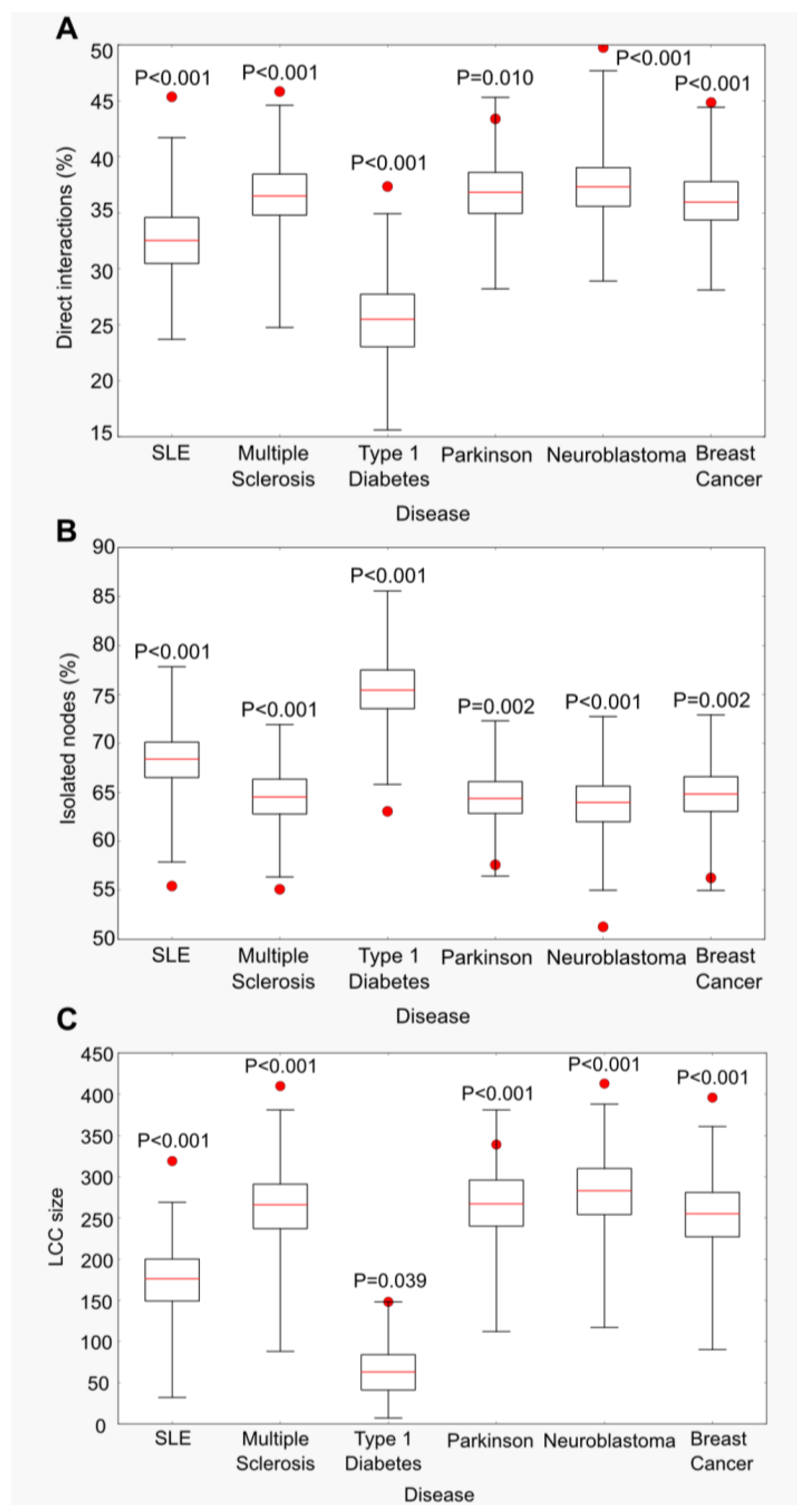
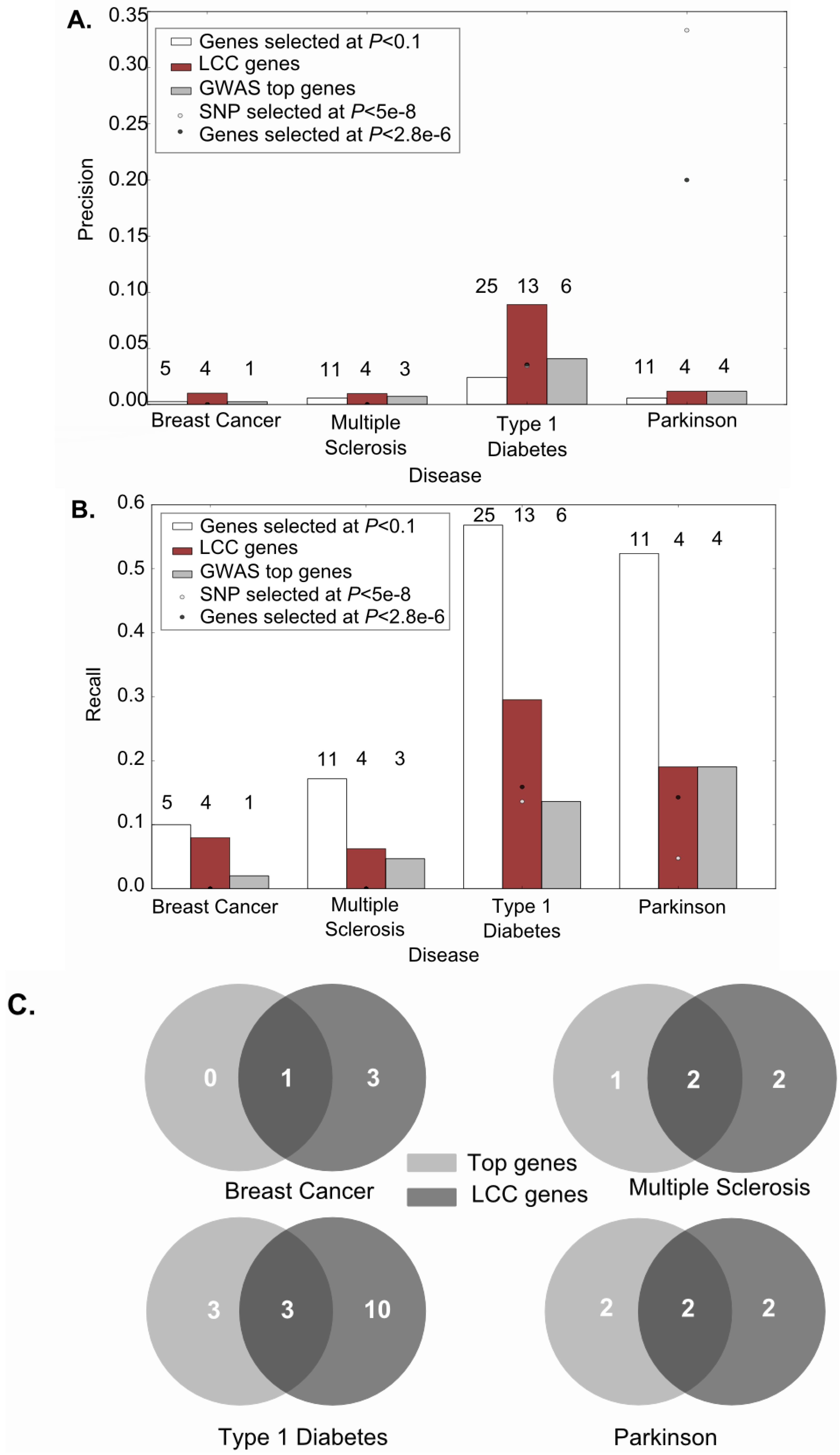
2.3. A Case Study: Breast Cancer Largest Connected Component (LCC) Genes Were Supported by Multiple Sources of Experimental Data
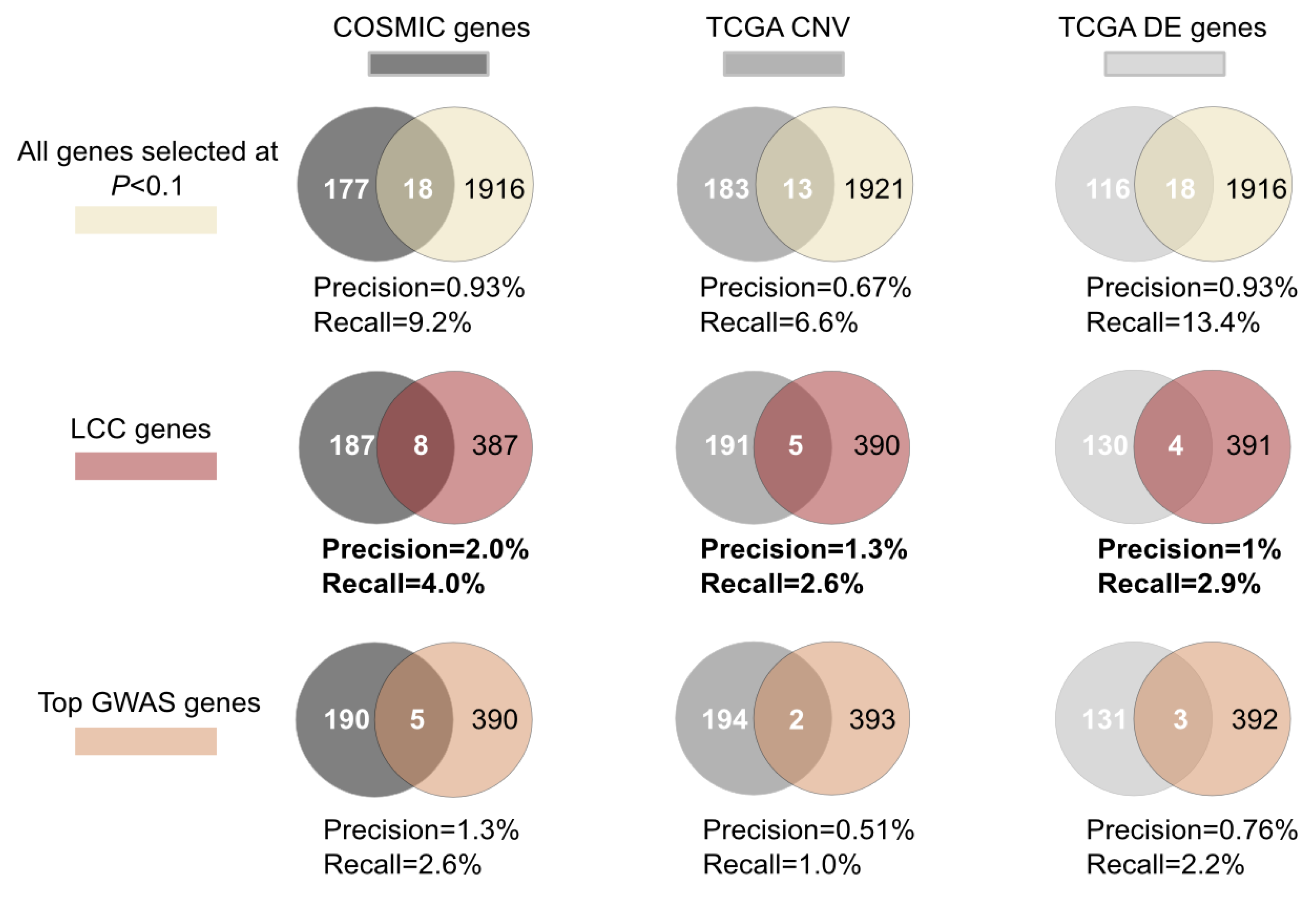
2.4. Novel Breast Cancer Susceptibility Genes
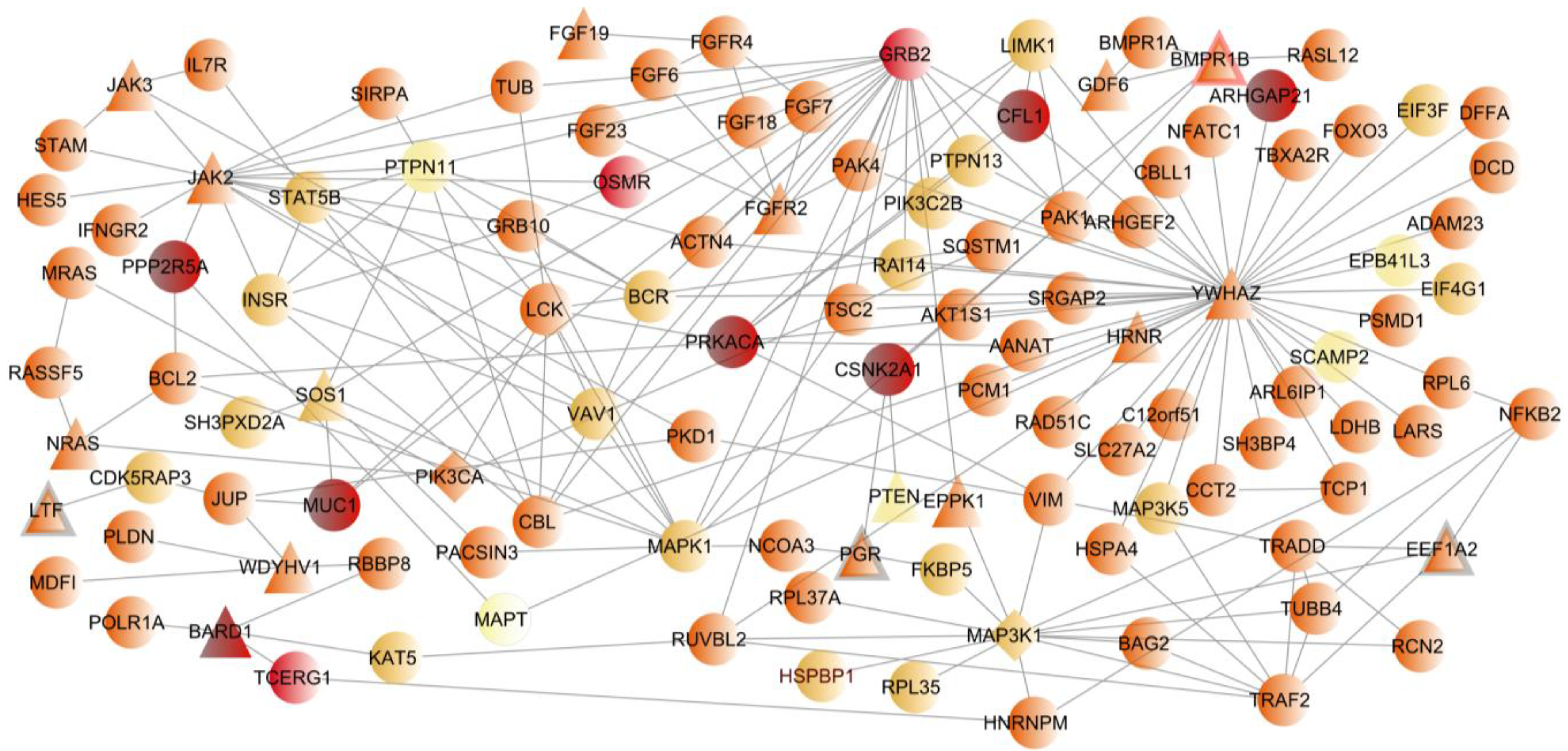
3. Discussion
4. Experimental Section
4.1. GWAS Datasets
4.2. Integration of Gene Association Data with Protein-Protein Interaction Data
4.3. PPI Network Analysis
4.4. Performance against Benchmarks
4.5. Case Study: Breast Cancer
5. Conclusions
Supplementary Materials
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- The Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3000 shared controls. Nature 2007, 447, 661–678. [Google Scholar]
- Bradfield, J.P.; Qu, H.-Q.; Wang, K.; Zhang, H.; Sleiman, P.M.; Kim, C.E.; Mentch, F.D.; Qiu, H.; Glessner, J.T.; Thomas, K.A.; et al. A genome-wide meta-analysis of six type 1 diabetes cohorts identifies multiple associated loci. PLoS Genet. 2011, 7, e1002293. [Google Scholar] [PubMed]
- Franke, A.; McGovern, D.P.B.; Barrett, J.C.; Wang, K.; Radford-Smith, G.L.; Ahmad, T.; Lees, C.W.; Balschun, T.; Lee, J.; Roberts, R.; et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat. Genet. 2010, 42, 1118–1125. [Google Scholar] [PubMed]
- Lango Allen, H.; Estrada, K.; Lettre, G.; Berndt, S.I.; Weedon, M.N.; Rivadeneira, F.; Willer, C.J.; Jackson, A.U.; Vedantam, S.; Raychaudhuri, S.; et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 2010, 467, 832–838. [Google Scholar] [PubMed]
- Manolio, T.A.; Collins, F.S.; Cox, N.J.; Goldstein, D.B.; Hindorff, L.A.; Hunter, D.J.; McCarthy, M.I.; Ramos, E.M.; Cardon, L.R.; Chakravarti, A.; et al. Finding the missing heritability of complex diseases. Nature 2009, 461, 747–753. [Google Scholar] [PubMed]
- Dudbridge, F.; Gusnanto, A. Estimation of significance thresholds for genomewide association scans. Genet. Epidemiol. 2008, 32, 227–234. [Google Scholar]
- Risch, N.; Merikangas, K. The future of genetic studies of complex human diseases. Science 1996, 273, 1516–1517. [Google Scholar] [PubMed]
- Yang, J.; Benyamin, B.; McEvoy, B.P.; Gordon, S.; Henders, A.K.; Nyholt, D.R.; Madden, P.A.; Heath, A.C.; Martin, N.G.; Montgomery, G.W.; et al. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 2010, 42, 565–569. [Google Scholar] [PubMed]
- Purcell, S.M.; Wray, N.R.; Stone, J.L.; Visscher, P.M.; O’Donovan, M.C.; Sullivan, P.F.; Sklar, P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 2009, 460, 748–752. [Google Scholar] [PubMed]
- Gargalovic, P.S.; Imura, M.; Zhang, B.; Gharavi, N.M.; Clark, M.J.; Pagnon, J.; Yang, W.-P.; He, A.; Truong, A.; Patel, S.; et al. Identification of inflammatory gene modules based on variations of human endothelial cell responses to oxidized lipids. PNAS 2006, 103, 12741–12746. [Google Scholar] [PubMed]
- Horvath, S.; Zhang, B.; Carlson, M.; Lu, K.V.; Zhu, S.; Felciano, R.M.; Laurance, M.F.; Zhao, W.; Qi, S.; Chen, Z.; et al. Analysis of oncogenic signaling networks in glioblastoma identifies ASPM as a molecular target. Proc. Natl. Acad. Sci. USA 2006, 103, 17402–17407. [Google Scholar] [PubMed]
- Presson, A.P.; Sobel, E.M.; Papp, J.C.; Suarez, C.J.; Whistler, T.; Rajeevan, M.S.; Vernon, S.D.; Horvath, S. Integrated weighted gene co-expression network analysis with an application to chronic fatigue syndrome. BMC Syst. Biol. 2008, 2, 95. [Google Scholar] [PubMed]
- Torkamani, A.; Schork, N.J. Identification of rare cancer driver mutations by network reconstruction. Genome Res. 2009, 19, 1570–1578. [Google Scholar]
- Voineagu, I.; Wang, X.; Johnston, P.; Lowe, J.K.; Tian, Y.; Horvath, S.; Mill, J.; Cantor, R.M.; Blencowe, B.J.; Geschwind, D.H. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature 2011, 474, 380–384. [Google Scholar] [PubMed]
- Akula, N.; Baranova, A.; Seto, D.; Solka, J.; Nalls, M.A.; Singleton, A.; Ferrucci, L.; Tanaka, T.; Bandinelli, S.; Cho, Y.S.; et al. A network-based approach to prioritize results from genome-wide association studies. PLoS One 2011, 6, e24220. [Google Scholar] [PubMed]
- Baranzini, S.E.; Galwey, N.W.; Wang, J.; Khankhanian, P.; Lindberg, R.; Pelletier, D.; Wu, W.; Uitdehaag, B.M.; Kappos, L.; Polman, C.H.; et al. Pathway and network-based analysis of genome-wide association studies in multiple sclerosis. Hum. Mol. Genet. 2009, 18, 2078–2090. [Google Scholar] [PubMed]
- Calvano, S.E.; Xiao, W.; Richards, D.R.; Felciano, R.M.; Baker, H.V.; Cho, R.J.; Chen, R.O.; Brownstein, B.H.; Cobb, J.P.; Tschoeke, S.K.; et al. A network-based analysis of systemic inflammation in humans. Nature 2005, 437, 1032–1037. [Google Scholar] [PubMed]
- Hwang, D.; Lee, I.Y.; Yoo, H.; Gehlenborg, N.; Cho, J.-H.; Petritis, B.; Baxter, D.; Pitstick, R.; Young, R.; Spicer, D.; et al. A systems approach to prion disease. Mol. Syst. Biol. 2009, 5, 252. [Google Scholar] [PubMed]
- Jensen, M.K.; Pers, T.H.; Dworzynski, P.; Girman, C.J.; Brunak, S.; Rimm, E.B. Protein interaction-based genome-wide analysis of incident coronary heart disease. Circ. Cardiovasc. Genet. 2011, 4, 549–556. [Google Scholar]
- Jia, P.; Zheng, S.; Long, J.; Zheng, W.; Zhao, Z. dmGWAS: Dense module searching for genome-wide association studies in protein-protein interaction networks. Bioinformatics 2011, 27, 95–102. [Google Scholar]
- Lee, E.; Jung, H.; Radivojac, P.; Kim, J.-W.; Lee, D. Analysis of AML genes in dysregulated molecular networks. BMC Bioinform. 2009, 10, S2. [Google Scholar]
- Lee, I.; Blom, U.M.; Wang, P.I.; Shim, J.E.; Marcotte, E.M. Prioritizing candidate disease genes by network-based boosting of genome-wide association data. Genome Res. 2011, 21, 1109–1121. [Google Scholar] [PubMed]
- Liu, M.; Liberzon, A.; Kong, S.W.; Lai, W.R.; Park, P.J.; Kohane, I.S.; Kasif, S. Network-based analysis of affected biological processes in type 2 diabetes models. PLoS Genet. 2007, 3, e96. [Google Scholar] [PubMed]
- Nibbe, R.K.; Koyutürk, M.; Chance, M.R. An integrative-omics approach to identify functional sub-networks in human colorectal cancer. PLoS Comput. Biol. 2010, 6, e1000639. [Google Scholar]
- Cantor, R.M.; Lange, K.; Sinsheimer, J.S. Prioritizing GWAS results: A review of statistical methods and recommendations for their application. Am. J. Hum. Genet. 2010, 86, 6–22. [Google Scholar]
- Elbers, C.C.; van Eijk, K.R.; Franke, L.; Mulder, F.; van der Schouw, Y.T.; Wijmenga, C.; Onland-Moret, N.C. Using genome-wide pathway analysis to unravel the etiology of complex diseases. Genet. Epidemiol. 2009, 33, 419–431. [Google Scholar] [PubMed]
- Hirschhorn, J.N. Genomewide association studies—Illuminating biologic pathways. N. Engl. J. Med. 2009, 360, 1699–1701. [Google Scholar] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. PNAS 2005, 102, 15545–15550. [Google Scholar] [PubMed]
- Torkamani, A.; Topol, E.J.; Schork, N.J. Pathway analysis of seven common diseases assessed by genome-wide association. Genomics 2008, 92, 265–272. [Google Scholar] [PubMed]
- Pujana, M.A.; Han, J.D.; Starita, L.M.; Stevens, K.N.; Tewari, M.; Ahn, J.S.; Rennert, G.; Moreno, V.; Kirchhoff, T.; Gold, B.; et al. Network modeling links breast cancer susceptibility and centrosome dysfunction. Nat. Genet. 2007, 39, 1338–1349. [Google Scholar] [PubMed]
- Chuang, H.Y.; Lee, E.; Liu, Y.T.; Lee, D.; Ideker, T. Network-based classification of breast cancer metastasis. Mol. Syst. Biol. 2007, 3, 140. [Google Scholar] [PubMed]
- Goh, K.I.; Cusick, M.E.; Valle, D.; Childs, B.; Vidal, M.; Barabasi, A.L. The human disease network. Proc. Natl. Acad. Sci. USA 2007, 104, 8685–8690. [Google Scholar] [PubMed]
- Lage, K.; Karlberg, E.O.; Størling, Z.M.; Olason, P.I.; Pedersen, A.G.; Rigina, O.; Hinsby, A.M.; Tümer, Z.; Pociot, F.; Tommerup, N.; et al. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat. Biotechnol. 2007, 25, 309–316. [Google Scholar] [PubMed]
- Oti, M.; Brunner, H.G. The modular nature of genetic diseases. Clin. Genet. 2007, 71, 1–11. [Google Scholar]
- Goehler, H.; Lalowski, M.; Stelzl, U.; Waelter, S.; Stroedicke, M.; Worm, U.; Droege, A.; Lindenberg, K.S.; Knoblich, M.; Haenig, C.; et al. A protein interaction network links GIT1, an enhancer of huntingtin aggregation, to Huntington’s disease. Mol. Cell 2004, 15, 853–865. [Google Scholar] [PubMed]
- Lim, J.; Hao, T.; Shaw, C.; Patel, A.J.; Szabo, G.; Rual, J.F.; Fisk, C.J.; Li, N.; Smolyar, A.; Hill, D.E.; et al. A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell 2006, 125, 801–814. [Google Scholar] [PubMed]
- Rossin, E.J.; Lage, K.; Raychaudhuri, S.; Xavier, R.J.; Tatar, D.; Benita, Y.; Cotsapas, C.; Daly, M.J.; Constortium, I.I.B.D.G. Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS Genet. 2011, 7, e1001273. [Google Scholar] [PubMed]
- Bergholdt, R.; Storling, Z.M.; Lage, K.; Karlberg, E.O.; Olason, P.I.; Aalund, M.; Nerup, J.; Brunak, S.; Workman, C.T.; Pociot, F. Integrative analysis for finding genes and networks involved in diabetes and other complex diseases. Genome Biol. 2007, 8, R253. [Google Scholar] [PubMed]
- Burren, O.S.; Adlem, E.C.; Achuthan, P.; Christensen, M.; Coulson, R.M.R.; Todd, J.A. T1DBase: Update 2011, organization and presentation of large-scale data sets for type 1 diabetes research. Nucleic Acids Res. 2011, 39, D997–D1001. [Google Scholar] [PubMed]
- Kupershmidt, I.; Su, Q.J.; Grewal, A.; Sundaresh, S.; Halperin, I.; Flynn, J.; Shekar, M.; Wang, H.; Park, J.; Cui, W.; et al. Ontology-based meta-analysis of global collections of high-throughput public data. PLoS One 2010, 5, e13066. [Google Scholar] [PubMed]
- Lill, C.M.; Roehr, J.T.; McQueen, M.B.; Bagade, S.; Schjeide, B.M.; Zipp, F.; Bertram, L. The MSGene database. Alzheimer Research Forum. Available online: http://www.msgene.org/ (accessed on 27 March 2012).
- Lill, C.M.; Roehr, J.T.; McQueen, M.B.; Kavvoura, F.K.; Bagade, S.; Schjeide, B.-M.M.; Schjeide, L.M.; Meissner, E.; Zauft, U.; Allen, N.C.; et al. Comprehensive research synopsis and systematic meta-analyses in Parkinson’s’s disease genetics: The PDGene database. PLoS Genet. 2012, 8, e1002548. [Google Scholar]
- Forbes, S.A.; Bhamra, G.; Bamford, S.; Dawson, E.; Kok, C.; Clements, J.; Menzies, A.; Teague, J.W.; Futreal, P.A.; Stratton, M.R. The catalogue of somatic mutations in cancer (COSMIC). Curr. Protoc. Hum. Genet. 2008. [Google Scholar] [CrossRef]
- Forbes, S.A.; Bindal, N.; Bamford, S.; Cole, C.; Kok, C.Y.; Beare, D.; Jia, M.; Shepherd, R.; Leung, K.; Menzies, A.; et al. COSMIC: Mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2010, 39, D945–D950. [Google Scholar] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [PubMed]
- Futreal, P.A.; Coin, L.; Marshall, M.; Down, T.; Hubbard, T.; Wooster, R.; Rahman, N.; Stratton, M.R. A census of human cancer genes. Nat. Rev. Cancer 2004, 4, 177–183. [Google Scholar] [PubMed]
- Carles, A.; Millon, R.; Cromer, A.; Ganguli, G.; Lemaire, F.; Young, J.; Wasylyk, C.; Muller, D.; Schultz, I.; Rabouel, Y.; et al. Head and neck squamous cell carcinoma transcriptome analysis by comprehensive validated differential display. Oncogene 2006, 25, 1821–1831. [Google Scholar] [PubMed]
- Kan, B.; Esapa, C.; Sipahi, T.; Nacar, C.; Ozer, F.; Sayhan, N.B.; Kaynar, M.Y.; Sarioglu, A.C.; Harris, P.E. G protein mutations in pituitary tumors: A study on Turkish patients. Pituitary 2003, 6, 75–80. [Google Scholar] [PubMed]
- Lascorz, J.; Försti, A.; Chen, B.; Buch, S.; Steinke, V.; Rahner, N.; Holinski-Feder, E.; Morak, M.; Schackert, H.K.; Görgens, H.; et al. Genome-wide association study for colorectal cancer identifies risk polymorphisms in German familial cases and implicates MAPK signalling pathways in disease susceptibility. Carcinogenesis 2010, 31, 1612–1619. [Google Scholar] [PubMed]
- O. charoenrat, P.; Rusch, V.; Talbot, S.G.; Sarkaria, I.; Viale, A.; Socci, N.; Ngai, I.; Rao, P.; Singh, B. Casein kinase II α subunit and C1-inhibitor are independent predictors of outcome in patients with squamous cell carcinoma of the lung. Clin. Cancer Res. 2004, 10, 5792–5803. [Google Scholar] [CrossRef] [PubMed]
- Ideker, T.; Ozier, O.; Schwikowski, B.; Siegel, A.F. Discovering regulatory and signalling circuits in molecular interaction networks. Bioinformatics 2002, 18, S233–S240. [Google Scholar] [PubMed]
- Guney, E.; Oliva, B. Exploiting protein-protein interaction networks for genome-wide disease-gene prioritization. PLoS One 2012, 7, e43557. [Google Scholar] [PubMed]
- Hong, M.-G.; Pawitan, Y.; Magnusson, P.K.E.; Prince, J.A. Strategies and issues in the detection of pathway enrichment in genome-wide association studies. Hum. Genet. 2009, 126, 289–301. [Google Scholar] [PubMed]
- Lehne, B.; Lewis, C.M.; Schlitt, T. From SNPs to genes: Disease association at the gene level. PLoS One 2011, 6, e20133. [Google Scholar] [PubMed]
- Wang, K.; Li, M.; Bucan, M. Pathway-based approaches for analysis of genomewide association studies. Am. J. Hum. Genet. 2007, 81, 1278–1283. [Google Scholar]
- Segre, A.V.; Groop, L.; Mootha, V.K.; Daly, M.J.; Altshuler, D. Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet. 2010, 6, e1001058. [Google Scholar] [PubMed]
- Liu, S.; Zhang, C.; Zhou, Y. Uneven size distribution of mammalian genes in the number of tissues expressed and in the number of co-expressed genes. Hum. Mol. Genet. 2006, 15, 1313–1318. [Google Scholar] [PubMed]
- Consortium, T.E.P. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar] [PubMed]
- De Brakeleer, S.; de Greve, J.; Loris, R.; Janin, N.; Lissens, W.; Sermijn, E.; Teugels, E. Cancer predisposing missense and protein truncating BARD1 mutations in non-BRCA1 or BRCA2 breast cancer families. Hum. Mutat. 2010, 31, E1175–E1185. [Google Scholar]
- Huo, X.; Hu, Z.; Zhai, X.; Wang, Y.; Wang, S.; Wang, X.; Qin, J.; Chen, W.; Jin, G.; Liu, J.; et al. Common non-synonymous polymorphisms in the BRCA1 Associated RING Domain (BARD1) gene are associated with breast cancer susceptibility: A case-control analysis. Breast Cancer Res. Treat. 2007, 102, 329–337. [Google Scholar] [PubMed]
- Leth-Larsen, R.; Terp, M.G.; Christensen, A.G.; Elias, D.; Kühlwein, T.; Jensen, O.N.; Petersen, O.W.; Ditzel, H.J. Functional heterogeneity within the CD44 high human breast cancer stem cell-like compartment reveals a gene signature predictive of distant metastasis. Mol. Med. 2012, 18, 1109–1121. [Google Scholar] [PubMed]
- Mukhopadhyay, P.; Chakraborty, S.; Ponnusamy, M.P.; Lakshmanan, I.; Jain, M.; Batra, S.K. Mucins in the pathogenesis of breast cancer: Implications in diagnosis, prognosis and therapy. Biochim. Biophys. Acta 2011, 1815, 224–240. [Google Scholar] [PubMed]
- Leong, S.; McKay, M.J.; Christopherson, R.I.; Baxter, R.C. Biomarkers of breast cancer apoptosis induced by chemotherapy and TRAIL. J. Proteome Res. 2012, 11, 1240–1250. [Google Scholar] [PubMed]
- Zhang, Y.; Tong, X. Expression of the actin-binding proteins indicates that cofilin and fascin are related to breast tumour size. J. Int. Med. Res. 2010, 38, 1042–1048. [Google Scholar] [PubMed]
- Dupont, W.D.; Breyer, J.P.; Bradley, K.M.; Schuyler, P.A.; Plummer, W.D.; Sanders, M.E.; Page, D.L.; Smith, J.R. Protein phosphatase 2A subunit gene haplotypes and proliferative breast disease modify breast cancer risk. Cancer 2010, 116, 8–19. [Google Scholar] [PubMed]
- Shih Ie, M.; Panuganti, P.K.; Kuo, K.T.; Mao, T.L.; Kuhn, E.; Jones, S.; Velculescu, V.E.; Kurman, R.J.; Wang, T.L. Somatic mutations of PPP2R1A in ovarian and uterine carcinomas. Am. J. Pathol. 2011, 178, 1442–1447. [Google Scholar] [PubMed]
- Baranzini, S.E.; Wang, J.; Gibson, R.A.; Galwey, N.; Naegelin, Y.; Barkhof, F.; Radue, E.W.; Lindberg, R.L.; Uitdehaag, B.M.; Johnson, M.R.; et al. Genome-wide association analysis of susceptibility and clinical phenotype in multiple sclerosis. Hum. Mol. Genet. 2009, 18, 767–778. [Google Scholar] [PubMed]
- Barrett, J.C.; Clayton, D.G.; Concannon, P.; Akolkar, B.; Cooper, J.D.; Erlich, H.A.; Julier, C.; Morahan, G.; Nerup, J.; Nierras, C.; et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat. Genet. 2009, 41, 703–707. [Google Scholar] [PubMed]
- Hom, G.; Graham, R.R.; Modrek, B.; Taylor, K.E.; Ortmann, W.; Garnier, S.; Lee, A.T.; Chung, S.A.; Ferreira, R.C.; Pant, P.V.; et al. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N. Engl. J. Med. 2008, 358, 900–909. [Google Scholar] [PubMed]
- Hunter, D.J.; Kraft, P.; Jacobs, K.B.; Cox, D.G.; Yeager, M.; Hankinson, S.E.; Wacholder, S.; Wang, Z.; Welch, R.; Hutchinson, A.; et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet. 2007, 39, 870–874. [Google Scholar] [PubMed]
- Maris, J.M.; Mosse, Y.P.; Bradfield, J.P.; Hou, C.; Monni, S.; Scott, R.H.; Asgharzadeh, S.; Attiyeh, E.F.; Diskin, S.J.; Laudenslager, M.; et al. Chromosome 6p22 locus associated with clinically aggressive neuroblastoma. N. Engl. J. Med. 2008, 358, 2585–2593. [Google Scholar] [PubMed]
- Simon-Sanchez, J.; Schulte, C.; Bras, J.M.; Sharma, M.; Gibbs, J.R.; Berg, D.; Paisan-Ruiz, C.; Lichtner, P.; Scholz, S.W.; Hernandez, D.G.; et al. Genome-wide association study reveals genetic risk underlying Parkinson’s’s disease. Nat. Genet. 2009, 41, 1308–1312. [Google Scholar] [PubMed]
- Bader, G.D.; Betel, D.; Hogue, C.W.V. BIND: The biomolecular interaction network database. Nucleic Acids Res. 2003, 31, 248–250. [Google Scholar] [PubMed]
- Ceol, A.; Chatr Aryamontri, A.; Licata, L.; Peluso, D.; Briganti, L.; Perfetto, L.; Castagnoli, L.; Cesareni, G. MINT, the molecular interaction database: 2009 update. Nucleic Acids Res. 2010, 38, D532–D539. [Google Scholar] [PubMed]
- Kerrien, S.; Aranda, B.; Breuza, L.; Bridge, A.; Broackes-Carter, F.; Chen, C.; Duesbury, M.; Dumousseau, M.; Feuermann, M.; Hinz, U.; et al. The IntAct molecular interaction database in 2012. Nucleic Acids Res. 2011, 40, D841–D846. [Google Scholar] [PubMed]
- Keshava Prasad, T.S.; Goel, R.; Kandasamy, K.; Keerthikumar, S.; Kumar, S.; Mathivanan, S.; Telikicherla, D.; Raju, R.; Shafreen, B.; Venugopal, A.; et al. Human protein reference database—2009 update. Nucleic Acids Res. 2009, 37, D767–D772. [Google Scholar]
- Mishra, G.R.; Suresh, M.; Kumaran, K.; Kannabiran, N.; Suresh, S.; Bala, P.; Shivakumar, K.; Anuradha, N.; Reddy, R.; Raghavan, T.M.; et al. Human protein reference database—2006 update. Nucleic Acids Res. 2006, 34, D411–414. [Google Scholar] [PubMed]
- Pagel, P.; Kovac, S.; Oesterheld, M.; Brauner, B.; Dunger-Kaltenbach, I.; Frishman, G.; Montrone, C.; Mark, P.; Stümpflen, V.; Mewes, H.-W.; et al. The MIPS mammalian protein-protein interaction database. Bioinformatics 2005, 21, 832–834. [Google Scholar] [PubMed]
- Peri, S.; Navarro, J.D.; Amanchy, R.; Kristiansen, T.Z.; Jonnalagadda, C.K.; Surendranath, V.; Niranjan, V.; Muthusamy, B.; Gandhi, T.K.B.; Gronborg, M.; et al. Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res. 2003, 13, 2363–2371. [Google Scholar]
- Stark, C. BioGRID: A general repository for interaction datasets. Nucleic Acids Res. 2006, 34, D535–D539. [Google Scholar] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [PubMed]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Correia, C.; Diekmann, Y.; Vicente, A.M.; Pereira-Leal, J.B. Hope for GWAS: Relevant Risk Genes Uncovered from GWAS Statistical Noise. Int. J. Mol. Sci. 2014, 15, 17601-17621. https://doi.org/10.3390/ijms151017601
Correia C, Diekmann Y, Vicente AM, Pereira-Leal JB. Hope for GWAS: Relevant Risk Genes Uncovered from GWAS Statistical Noise. International Journal of Molecular Sciences. 2014; 15(10):17601-17621. https://doi.org/10.3390/ijms151017601
Chicago/Turabian StyleCorreia, Catarina, Yoan Diekmann, Astrid M. Vicente, and José B. Pereira-Leal. 2014. "Hope for GWAS: Relevant Risk Genes Uncovered from GWAS Statistical Noise" International Journal of Molecular Sciences 15, no. 10: 17601-17621. https://doi.org/10.3390/ijms151017601



