Molecular Typing of Mastitis-Causing Staphylococcus aureus Isolated from Heifers and Cows
Abstract
:1. Introduction
2. Results
2.1. Isolation of S. aureus
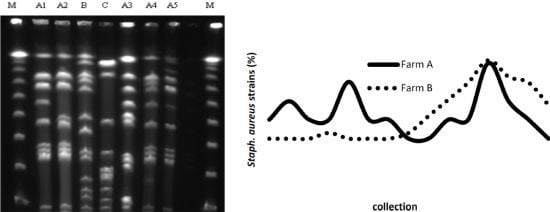
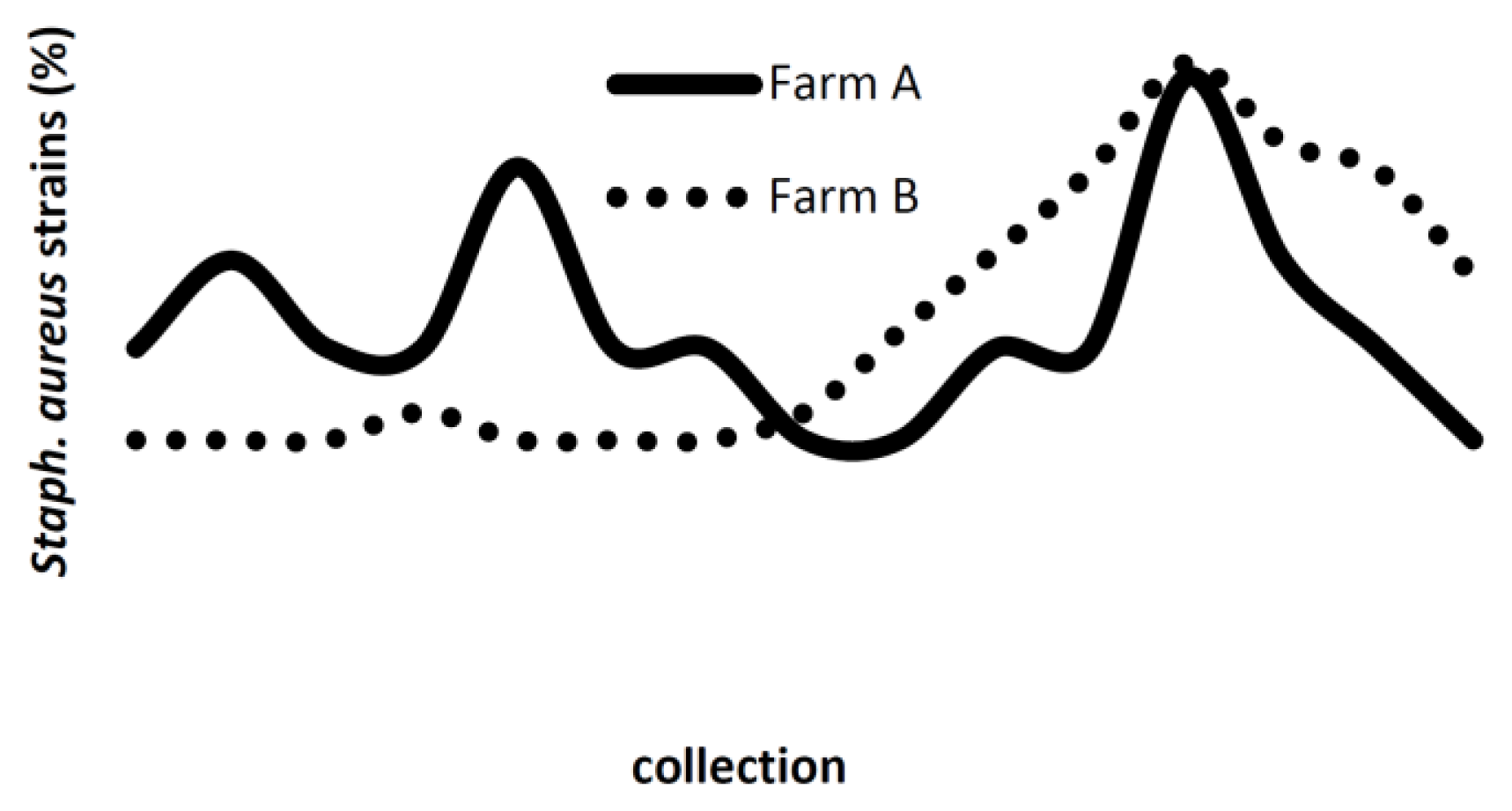
2.2. PFGE
3. Discussion
4. Materials and Methods
4.1. Collection of Biological Material
4.2. Isolation and Identification
4.3. Species Confirmation by PCR
4.4. PFGE
4.5. Statistical Analysis
5. Conclusions
Acknowledgments
Conflict of Interest
References
- De Vliegher, S.; Fox, L.K.; Piepers, S.; McDougall, S.; Barkema, H.W. Invited review: Mastitis in dairy heifers: Nature of the disease, potential impact, prevention, and control. J. Dairy Sci 2012, 95, 1025–1040. [Google Scholar]
- Capurro, A.; Aspán, A.; Unnerstad, H.E.; Waller, K.P.; Artursson, K. Identification of potential sources of Staphylococcus aureus in herds with mastitis problems. J. Dairy Sci 2010, 93, 180–191. [Google Scholar]
- Oliver, S.P.; Mitchell, B.A. Intramammary infections in primigravid heifers near parturition. J. Dairy Sci 1983, 66, 1180–1183. [Google Scholar]
- Nickerson, S.C.; Owens, W.E.; Boddie, R.L. Mastitis in dairy heifers: Initial studies on prevalence and control. J. Dairy Sci 1995, 78, 1607–1618. [Google Scholar]
- Piepers, S.; Opsomer, G.; Barkema, H.W.; de Kruif, A.; de Vliegher, S. Heifers infected with coagulase-negative staphylococci in early lactation have fewer cases of clinical mastitis and higher milk production in their first lactation than noinfected heifers. J. Dairy Sci 2010, 93, 2014–2024. [Google Scholar]
- Oliver, S.P.; Gillespie, B.E.; Headrick, S.J.; Lewis, M.J.; Dowlen, H.H. Prevalence, risk factors, and strategies for controlling mastitis in heifers during the periparturient period. Int. J. Appl. Res. Vet. Med 2005, 3, 150–162. [Google Scholar]
- Bannerman, T.L.; Hancock, G.A.; Tenover, F.C.; Miller, J.M. Pulsedfield gel electrophoresis as a replacement for bacteriophage typing of Staphylococcus aureus. J. Clin. Microbiol 1995, 33, 551–555. [Google Scholar]
- Pankey, J.W.; Drechsler, P.A.; Wildman, E.E. Mastitis prevalence in primigravid heifers at parturition. J. Dairy Sci 1991, 74, 1550–1552. [Google Scholar]
- Bava, L.; Zucali, M.; Brasca, M.; Zanini, L.; Sandrucci, A. Efficiency of cleaning procedure of milking equipment and bacterial quality of milk. Ital. J. Anim. Sci 2009, 8, 387–389. [Google Scholar]
- Smith, E.M.; Green, L.E.; Medley, G.F.; Bird, H.E.; Dowson, C.G. Multilocus sequence typing of Staphylococcus aureus isolated from high-somatic-cell-count cows and the environment of an organic dairy farm in the United Kigdom. J. Clin. Microbiol 2005, 43, 4731–4736. [Google Scholar]
- Roberson, J.R.; Fox, L.K.; Hancock, D.D.; Gay, J.M.; Besser, T.E. Coagulase-positive Staphylococcus intramammary infections in primiparous dairy cows. J. Dairy Sci 1994, 77, 958–969. [Google Scholar]
- Smith, T.H.; Fox, L.K.; Middleton, J.R. Outbreak of mastitis caused by one strain of Staphylococcus aureus in a closed dairy herd. J. Am. Vet. Med. Assoc 1998, 212, 553–556. [Google Scholar]
- Zadoks, R.; van Leeuwen, W.; Barkema, H.; Sampimon, O.; Verbrugh, H.; Schukken, Y.H.; van Belkum, A. Application of pulsed-field electrophoresis and binary typing as tools in veterinary clinical microbiology and molecular analysis of bovine and human Staphylococcus aureus isolates. Infect. Immun 2000, 38, 1931–1939. [Google Scholar]
- Akineden, O.; Annemuller, C.; Hassan, A.A.; LaMmler, C.; Wolter, W.; Zschock, M. Toxin genes and other characteristics of Staphylococcus aureus isolates from milk of cows with mastitis. Clin. Diagn. Lab. Immunol 2001, 8, 959–964. [Google Scholar]
- Smith, E.M.; Green, L.E.; Medley, G.F.; Bird, H.E.; Fox, L.K.; Schukken, Y.H.; Kruze, J.V.; Bradley, A.J.; Zadoks, R.N.; Dowson, C.G. Multilocus sequence typing of intercontinental bovine Staphylococcus aureus isolates. J. Clin. Microbiol 2005, 43, 4737–4743. [Google Scholar]
- Cabral, K.G.; Lämmler, C.; Zschöck, M.; Langoni, H.; de Sá, M.E.P.; Victória, C.; da Silva, A.V. Pheno and genotyping of Staphylococcus aureus, isolated from bovine milk samples from São Paulo State, Brazil. Can. J. Microbiol 2004, 50, 901–909. [Google Scholar]
- Su, C.; Herbelin, C.; Frieze, N.; Skardova, O.; Sordillo, M. Coagulase gene polymorphism of Staphylococcus aureus isolates from dairy cattle in different geographical areas. Epidemiol. Infect 1999, 122, 329–336. [Google Scholar]
- Harmon, R.J.; Eberhart, R.J.; Jasper, D.E.; Langlois, B.E.; Wilson, R.A. Microbiological Procedures for the Diagnosis of Bovine Udder Infection; National Mastitis Council, Inc.; Arlington, VA, USA, 1990; p. 34. [Google Scholar]
- APHA, Compendium of Methods for the Microbiological Examination of Foods, 4th ed; Dowes, F.P.; Ito, K. (Eds.) American Public Health Association: Washington, DC, USA, 2001.
- Martineau, F.; Picard, F.J.; Roy, P.H.; Ouellette, M.; Bergeron, M.G. Species-Specific and ubiquitous-DNA-based assays for rapid identification of Staphylococcus aureus. J. Clin. Microbiol 1998, 36, 618–623. [Google Scholar]
- McDougal, L.K.; Steward, C.D.; Killgore, G.E.; Chaitram, J.M.; McAllister, S.K.; Tenover, F.C. Pulsed-field gel electrophoresis typing of oxacilin-resistant Staphylococcus aureus isolates from the United: Establishing a national database. J. Clin. Microbiol 2003, 41, 5113–5120. [Google Scholar]
- Tenover, F.C.; Arbeit, R.D.; Goering, R.V.; Mickelsen, P.A.; Murray, B.E.; Persing, D.H.; Swaminathan, B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: Criteria for bacterial strain typing. J. Clin. Microbiol 1995, 33, 2233–2239. [Google Scholar]
- Dice, L.R. Measures of the amount of ecologic association between species. Ecology 1945, 26, 297–302. [Google Scholar]
- Sneath, P.H.A.; Sokal, R.R. Numerical Taxonomy-Principles and Practice of Numerical Classification; W. F. Freeman: San Francisco, CA, USA, 1973. [Google Scholar]
- Ward, J.H. Hierarchical grouping to optimize an objective function. J. Am. Stat. Assoc 1963, 58, 236–244. [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing; R Foundation for Statistical Computing: Vienna, Austria. Available online: http://www.r-project.org/ accessed on 22 August 2011.

| PFGE lineage | PFGE profile | Farm A | Farm B | Total | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Heifers | Cows | Heifers | Cows | ||||||||
| AF | RF | AF | RF | AF | RF | AF | RF | AF | RF | ||
| LA | A1 | - | - | - | - | 57 | 89.1 | 16 | 88.9 | 73 | 66.4 |
| A2 | 9 | 47.4 | 9 | 100.0 | 4 | 6.3 | - | - | 22 | 20.0 | |
| A3 | - | - | - | - | 1 | 1.6 | - | - | 1 | 0.9 | |
| A4 | - | - | - | - | - | - | 1 | 5.6 | 1 | 0.9 | |
| A5 | - | - | - | - | 1 | 1.6 | - | - | 1 | 0.9 | |
| LB | B1 | 10 | 52.6 | - | - | - | - | 1 | 5.6 | 11 | 10.0 |
| LC | C1 | - | - | - | - | 1 | 1.6 | - | - | 1 | 0.9 |
| Total | 19 | 100 | 9 | 100 | 64 | 100 | 18 | 100 | 110 | 100 | |
© 2013 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Castelani, L.; Santos, A.F.S.; Dos Santos Miranda, M.; Zafalon, L.F.; Pozzi, C.R.; Arcaro, J.R.P. Molecular Typing of Mastitis-Causing Staphylococcus aureus Isolated from Heifers and Cows. Int. J. Mol. Sci. 2013, 14, 4326-4333. https://doi.org/10.3390/ijms14024326
Castelani L, Santos AFS, Dos Santos Miranda M, Zafalon LF, Pozzi CR, Arcaro JRP. Molecular Typing of Mastitis-Causing Staphylococcus aureus Isolated from Heifers and Cows. International Journal of Molecular Sciences. 2013; 14(2):4326-4333. https://doi.org/10.3390/ijms14024326
Chicago/Turabian StyleCastelani, Lívia, Aline Franciele Silva Santos, Mariana Dos Santos Miranda, Luiz Francisco Zafalon, Claudia Rodrigues Pozzi, and Juliana Rodrigues Pozzi Arcaro. 2013. "Molecular Typing of Mastitis-Causing Staphylococcus aureus Isolated from Heifers and Cows" International Journal of Molecular Sciences 14, no. 2: 4326-4333. https://doi.org/10.3390/ijms14024326