Acidic Residue Glu199 Increases SUMOylation Level of Nuclear Hormone Receptor NR5A1
Abstract
:1. Introduction
2. Results
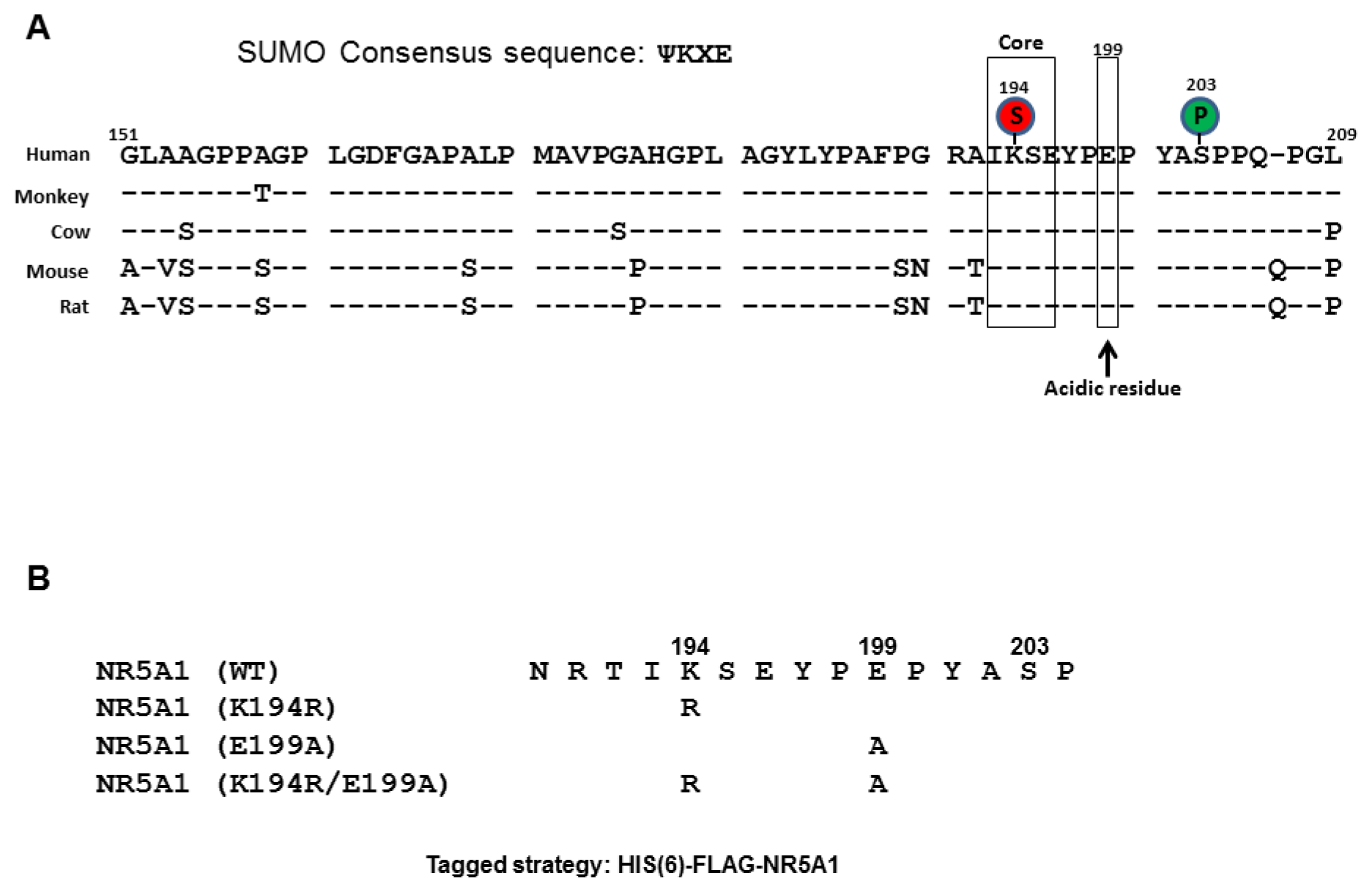
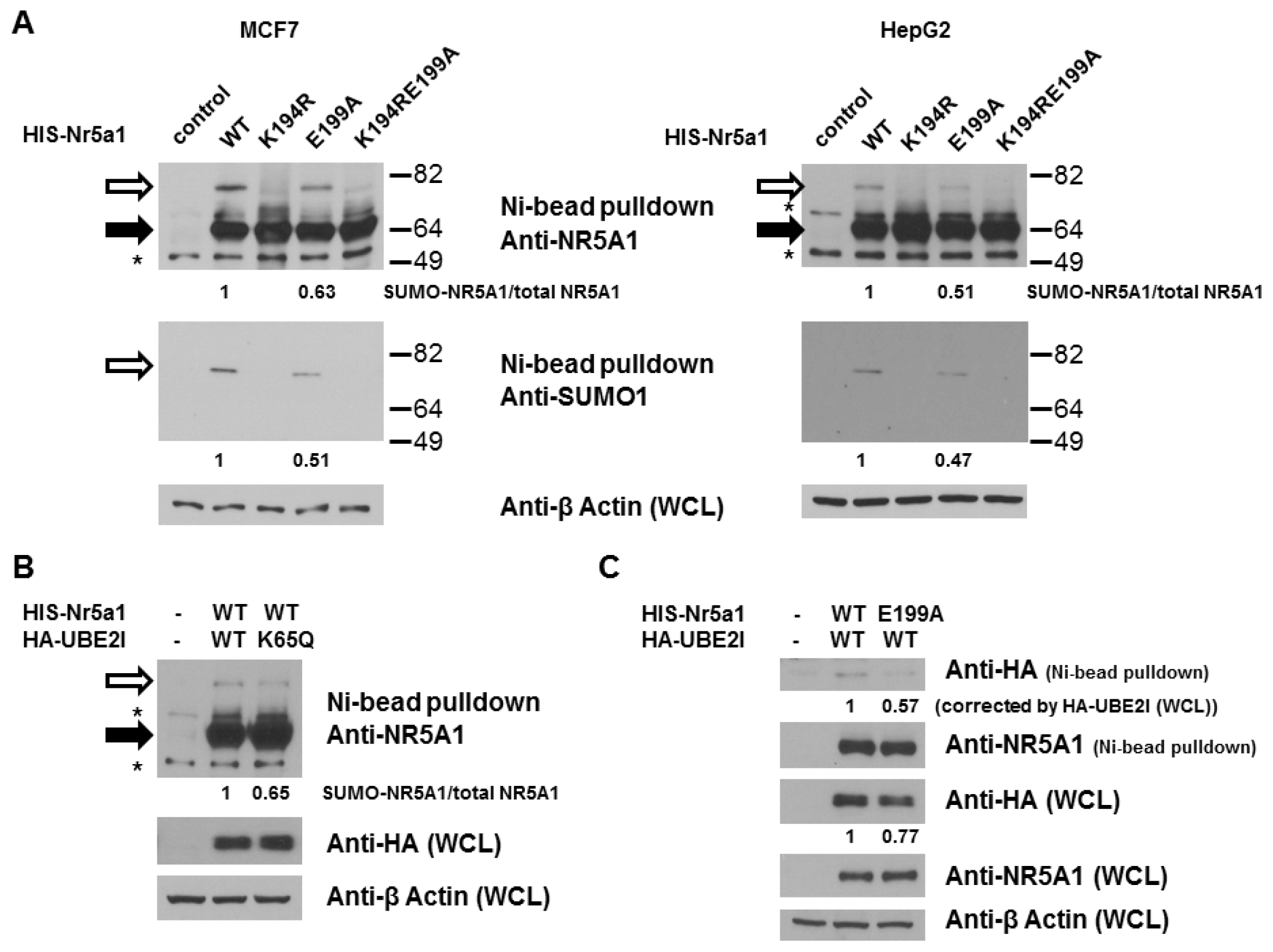
2.1. E199A Reduces NR5A1 SUMOylation
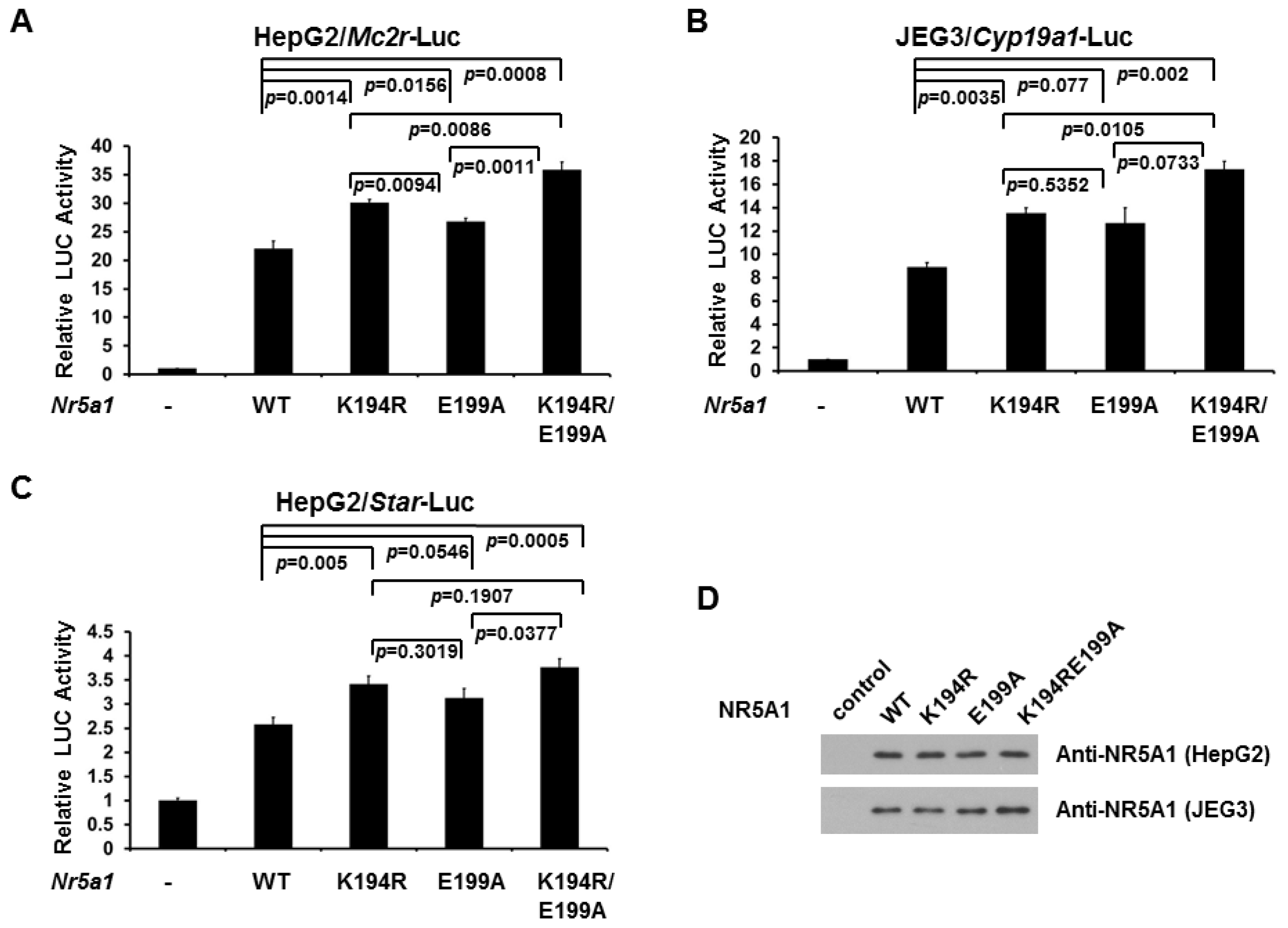
2.2. E199A Increases NR5A1 Transcriptional Activity
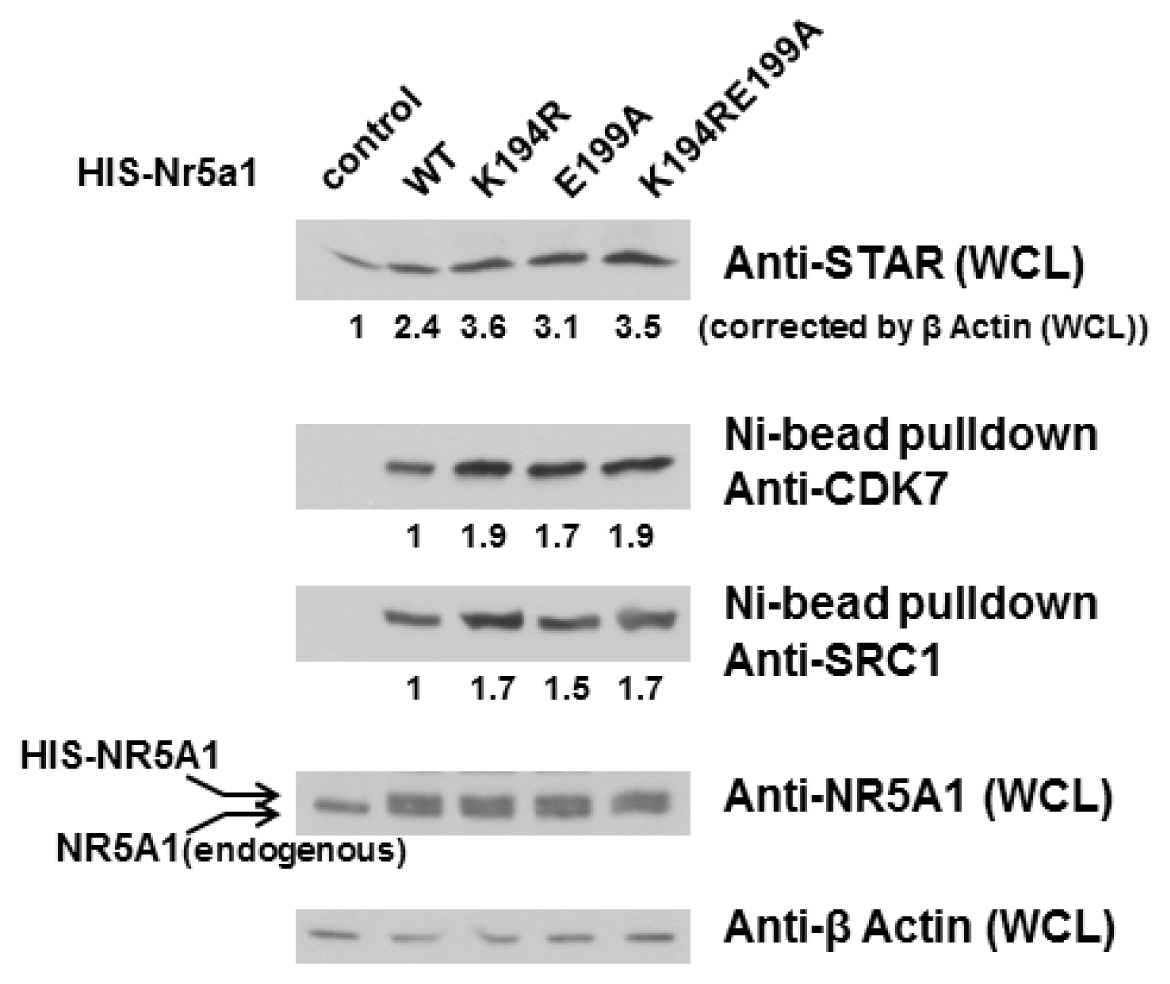
2.3. E199A Increases NR5A1’s Interaction with CDK7 and SRC1 as well as STAR Protein Level
3. Discussion
4. Experimental Section
4.1. Reagents
4.2. DNA Constructs
4.3. Cell Culture and Transfection
4.4. Immunoprecipitation Assay
4.5. Immunoblotting
4.6. In Vivo SUMOylation Assays
4.7. Statistical Analysis
5. Conclusions
Acknowledgments
Abbreviations
| AMH | muellerian-inhibiting factor |
| CDK7 | cyclin-dependent kinase 7 |
| CYP17A1 | steroid 17-alpha-hydroxylase/17,20-lyase |
| CYP19A1 | cytochrome p450 19A1 |
| DDX20 | ATP-dependent RNA helicase DDX20 |
| DGKθ | diacylglycerol kinase theta |
| EGR1 | early growth response protein 1 |
| GATA4 | transcription factor GATA-4 |
| GnRHR | gonadotropin-releasing hormone receptor |
| GSTA3 | glutathione S-tranferase A3 |
| MC2R | adrenocorticotropic hormone receptor |
| NCOR1 | nuclear receptor corepressor 1 |
| NDSM | negatively charged amino acid-dependent SUMOylation motif |
| NR0B1/DAX1 | nuclear receptor subfamily 0 group B member 1 |
| NR5A1 | steroidogenic factor-1 |
| PDSM | phosphorylation-dependent SUMOylation motif |
| PITX1 | pituitary homeobox 1 |
| SOX9 | transcription factor SOX-9 |
| SRC1/NCOA1 | nuclear receptor coactivator 1 |
| SREBP1 | sterol regulatory element-binding protein 1 |
| STAR | steroidogenic acute regulatory protein, mitochondrial |
| SUMO | small ubiquitin-like modifier |
| TIF2 | nuclear receptor coactivator 2 |
| UBE2I | SUMO-conjugating enzyme UBC9 |
| WT1 | Wilms tumor protein. |
Conflicts of Interest
References
- Shen, W.H.; Moore, C.C.; Ikeda, Y.; Parker, K.L.; Ingraham, H.A. Nuclear receptor steroidogenic factor 1 regulates the müllerian inhibiting substance gene: A link to the sex determination cascade. Cell 1994, 77, 651–661. [Google Scholar]
- Luo, X.; Ikeda, Y.; Parker, K.L. A cell-specific nuclear receptor is essential for adrenal and gonadal development and sexual differentiation. Cell 1994, 77, 481–490. [Google Scholar]
- Hammer, G.D.; Ingraham, H.A. Steroidogenic factor-1: Its role in endocrine organ development and differentiation. Front. Neuroendocrinol 1999, 20, 199–223. [Google Scholar]
- Val, P.; Lefrançois-Martinez, A.M.; Veyssière, G.; Martinez, A. SF-1 a key player in the development and differentiation of steroidogenic tissues. Nucl. Recept 2003, 1, 8. [Google Scholar]
- De Santa Barbara, P.; Bonneaud, N.; Boizet, B.; Desclozeaux, M.; Moniot, B.; Sudbeck, P.; Scherer, G.; Poulat, F.; Berta, P. Direct interaction of SRY-related protein SOX9 and steroidogenic factor 1 regulates transcription of the human anti-Müllerian hormone gene. Mol. Cell. Biol 1998, 18, 6653–6665. [Google Scholar]
- Dammer, E.B.; Leon, A.; Sewer, M.B. Coregulator exchange and sphingosine-sensitive cooperativity of steroidogenic factor-1, general control nonderepressed 5, p54, and p160 coactivators regulate cyclic adenosine 3′,5′-monophosphate-dependent cytochrome P450c17 transcription rate. Mol. Endocrinol 2007, 21, 415–438. [Google Scholar]
- Carlone, D.L.; Richards, J.S. Functional interactions, phosphorylation, and levels of 3′,5′-cyclic adenosine monophosphate-regulatory element binding protein and steroidogenic factor-1 mediate hormone-regulated and constitutive expression of aromatase in gonadal cells. Mol. Endocrinol 1997, 11, 292–304. [Google Scholar]
- Li, D.; Urs, A.N.; Allegood, J.; Leon, A.; Merrill, A.H., Jr.; Sewer, M.B. Cyclic AMP-stimulated interaction between steroidogenic factor 1 and diacylglycerol kinase theta facilitates induction of CYP17. Mol. Cell. Biol 2007, 27, 6669–6685. [Google Scholar]
- Ngan, E.S.; Cheng, P.K.; Leung, P.C.; Chow, B.K. Steroidogenic factor-1 interacts with a gonadotrope-specific element within the first exon of the human gonadotropin-releasing hormone receptor gene to mediate gonadotrope-specific expression. Endocrinology 1999, 140, 2452–2462. [Google Scholar]
- Matsumura, T.; Imamichi, Y.; Mizutani, T.; Ju, Y.; Yazawa, T.; Kawabe, S.; Kanno, M.; Ayabe, T.; Katsumata, N.; Fukami, M.; et al. Human glutathione S-transferase A (GSTA) family genes are regulated by steroidogenic factor 1 (SF-1) and are involved in steroidogenesis. FASEB J 2013, 27, 3198–3208. [Google Scholar]
- Winnay, J.N.; Hammer, G.D. Adrenocorticotropic hormone-mediated signaling cascades coordinate a cyclic pattern of steroidogenic factor 1-dependent transcriptional activation. Mol. Endocrinol 2006, 20, 147–166. [Google Scholar]
- Sugawara, T.; Kiriakidou, M.; McAllister, J.M.; Holt, J.A.; Arakane, F.; Strauss, J.F., III. Regulation of expression of the steroidogenic acute regulatory protein (StAR) gene: A central role for steroidogenic factor 1. Steroids 1997, 62, 5–9. [Google Scholar]
- Tremblay, J.J.; Marcil, A.; Gauthier, Y.; Drouin, J. Ptx1 regulates SF-1 activity by an interaction that mimics the role of the ligand-binding domain. EMBO J 1999, 18, 3431–3441. [Google Scholar]
- Tremblay, J.J.; Viger, R.S. Transcription factor GATA-4 enhances Müllerian inhibiting substance gene transcription through a direct interaction with the nuclear receptor SF-1. Mol. Endocrinol 1999, 13, 1388–1401. [Google Scholar]
- Dorn, C.; Ou, Q.; Svaren, J.; Crawford, P.A.; Sadovsky, Y. Activation of luteinizing hormone beta gene by gonadotropin-releasing hormone requires the synergy of early growth response-1 and steroidogenic factor-1. J. Biol. Chem 1999, 274, 13870–13876. [Google Scholar]
- Wilson, M.J.; Jeyasuria, P.; Parker, K.L.; Koopman, P. The transcription factors steroidogenic factor-1 and SOX9 regulate expression of Vanin-1 during mouse testis development. J. Biol. Chem 2005, 280, 5917–5923. [Google Scholar]
- Cai, K.; Sewer, M.B. cAMP-Stimulated transcription of diacylglycerol kinase theta (DGKθ) requires steroidogenic factor-1 (SF-1) and sterol responsive element binding protein 1 (SREBP1). J. Lipid Res 2013, 54, 2121–2132. [Google Scholar]
- Gurates, B.; Amsterdam, A.; Tamura, M.; Yang, S.; Zhou, J.; Fang, Z.; Amin, S.; Sebastian, S.; Bulun, S.E. WT1 and DAX-1 regulate SF-1-mediated human P450arom gene expression in gonadal cells. Mol. Cell. Endocrinol 2003, 208, 61–75. [Google Scholar]
- Ito, M.; Yu, R.N.; Jameson, J.L. Steroidogenic factor-1 contains a carboxy-terminal transcriptional activation domain that interacts with steroid receptor coactivator-1. Mol. Endocrinol 1998, 12, 290–301. [Google Scholar]
- Monté, D.; DeWitte, F.; Hum, D.W. Regulation of the human P450scc gene by steroidogenic factor 1 is mediated by CBP/p300. J. Biol. Chem 1998, 273, 4585–4591. [Google Scholar]
- Børud, B.; Hoang, T.; Bakke, M.; Jacob, A.L.; Lund, J.; Mellgren, G. The nuclear receptor coactivators p300/CBP/cointegrator-associated protein (p/CIP) and transcription intermediary factor 2 (TIF2) differentially regulate PKA-stimulated transcriptional activity of steroidogenic factor 1. Mol. Endocrinol 2002, 16, 757–773. [Google Scholar]
- Jordan, B.K.; Shen, J.H.; Olaso, R.; Ingraham, H.A.; Vilain, E. Wnt4 overexpression disrupts normal testicular vasculature and inhibits testosterone synthesis by repressing steroidogenic factor 1/beta-catenin synergy. Proc. Natl. Acad. Sci. USA 2003, 100, 10866–10871. [Google Scholar]
- Ou, Q.; Mouillet, J.F.; Yan, X.; Dorn, C.; Crawford, P.A.; Sadovsky, Y. The DEAD box protein DP103 is a regulator of steroidogenic factor-1. Mol. Endocrinol 2001, 15, 69–79. [Google Scholar]
- Crawford, P.A.; Dorn, C.; Sadovsky, Y.; Milbrandt, J. Nuclear receptor DAX-1 recruits nuclear receptor corepressor N-CoR to steroidogenic factor 1. Mol. Cell. Biol 1998, 18, 2949–2956. [Google Scholar]
- Xu, B.; Yang, W.H.; Gerin, I.; Hu, C.D.; Hammer, G.D.; Koenig, R.J. Dax-1 and steroid receptor RNA activator (SRA) function as transcriptional coactivators for steroidogenic factor 1 in steroidogenesis. Mol. Cell. Biol 2009, 29, 1719–1734. [Google Scholar]
- Urs, A.N.; Dammer, E.; Kelly, S.; Wang, E.; Merrill, A.H., Jr.; Sewer, M.B. Steroidogenic factor-1 is a sphingolipid binding protein. Mol. Cell. Endocrinol 2007, 265–266, 174–178. [Google Scholar]
- Shalitin, S.; Josefsberg, Z.; Vilain, E.; Shomrat, R.; Weintrob, N. Adrenal hypoplasia congenita with multiple pituitary hormone deficiency without documented mutation in DAX1 or SF1 gene. Mol. Genet. Metab 2002, 76, 157–161. [Google Scholar]
- Coutant, R.; Mallet, D.; Lahlou, N.; Bouhours-Nouet, N.; Guichet, A.; Coupris, L.; Croué, A.; Morel, Y. Heterozygous mutation of steroidogenic factor-1 in 46,XY subjects may mimic partial androgen insensitivity syndrome. J. Clin. Endocrinol. Metab 2007, 92, 2868–2873. [Google Scholar]
- Soardi, F.C.; Coeli, F.B.; Maciel-Guerra, A.T.; Guerra-Júnior, G.; Mello, M.P. Complete XY gonadal dysgenesis due to p.D293N homozygous mutation in the NR5A1 gene: A case study. J. Appl. Genet 2010, 51, 223–224. [Google Scholar]
- Köhler, B.; Lin, L.; Mazen, I.; Cetindag, C.; Biebermann, H.; Akkurt, I.; Rossi, R.; Hiort, O.; Grüters, A.; Achermann, J.C. The spectrum of phenotypes associated with mutations in steroidogenic factor 1 (SF-1, NR5A1, Ad4BP) includes severe penoscrotal hypospadias in 46,XY males without adrenal insufficiency. Eur. J. Endocrinol 2009, 161, 237–242. [Google Scholar]
- Philibert, P.; Zenaty, D.; Lin, L.; Soskin, S.; Audran, F.; Léger, J.; Achermann, J.C.; Sultan, C. Mutational analysis of steroidogenic factor 1 (NR5a1) in 24 boys with bilateral anorchia: A French collaborative study. Hum. Reprod 2007, 22, 3255–3261. [Google Scholar]
- Bashamboo, A.; Ferraz-de-Souza, B.; Lourenço, D.; Lin, L.; Sebire, N.J.; Montjean, D.; Bignon-Topalovic, J.; Mandelbaum, J.; Siffroi, J.P.; Christin-Maitre, S.; et al. Human male infertility associated with mutations in NR5A1 encoding steroidogenic factor 1. Am. J. Hum. Genet 2010, 87, 505–512. [Google Scholar]
- Druker, J.; Liberman, A.C.; Antunica-Noguerol, M.; Gerez, J.; Paez-Pereda, M.; Rein, T.; Iñiguez-Lluhí, J.A.; Holsboer, F.; Arzt, E. RSUME enhances glucocorticoid receptor SUMOylation and transcriptional activity. Mol. Cell. Biol 2013, 33, 2116–2127. [Google Scholar]
- Yang, F.; Yao, Y.; Jiang, Y.; Lu, L.; Ma, Y.; Dai, W. Sumoylation is important for stability, subcellular localization, and transcriptional activity of SALL4, an essential stem cell transcription factor. J. Biol. Chem 2012, 287, 38600–38608. [Google Scholar]
- Wang, C.M.; Brennan, V.C.; Gutierrez, N.M.; Wang, X.; Wang, L.; Yang, W.H. SUMOylation of ATF3 alters its transcriptional activity on regulation of TP53 gene. J. Cell. Biochem 2013, 114, 589–598. [Google Scholar]
- Lee, P.C.; Taylor-Jaffe, K.M.; Nordin, K.M.; Prasad, M.S.; Lander, R.M.; LaBonne, C. SUMOylated SoxE factors recruit Grg4 and function as transcriptional repressors in the neural crest. J. Cell. Biol 2012, 198, 799–813. [Google Scholar]
- Yang, W.H.; Heaton, J.H.; Brevig, H.; Mukherjee, S.; Iñiguez-Lluhí, J.A.; Hammer, G.D. SUMOylation inhibits SF-1 activity by reducing CDK7-mediated serine 203 phosphorylation. Mol. Cell. Biol 2009, 29, 613–625. [Google Scholar]
- Saitoh, H.; Hinchey, J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem 2000, 275, 6252–6258. [Google Scholar]
- Tatham, M.H.; Jaffray, E.; Vaughan, O.A.; Desterro, J.M.; Botting, C.H.; Naismith, J.H.; Hay, R.T. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem 2001, 276, 35368–35374. [Google Scholar]
- Noso, S.; Ikegami, H.; Fujisawa, T.; Kawabata, Y.; Asano, K.; Hiromine, Y.; Tsurumaru, M.; Sugihara, S.; Lee, I.; Kawasaki, E.; et al. Genetic heterogeneity in association of the SUMO4 M55V variant with susceptibility to type 1 diabetes. Diabetes 2005, 54, 3582–3586. [Google Scholar]
- Gareau, J.R.; Lima, C.D. The SUMO pathway: Emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell. Biol 2010, 11, 861–871. [Google Scholar]
- Wilkinson, K.A.; Henley, J.M. Mechanisms, regulation and consequences of protein SUMOylation. Biochem. J 2010, 428, 133–145. [Google Scholar]
- Abed, M.; Barry, K.C.; Kenyagin, D.; Koltun, B.; Phippen, T.M.; Delrow, J.J.; Parkhurst, S.M.; Orian, A. Degringolade, a SUMO-targeted ubiquitin ligase, inhibits Hairy/Groucho-mediated repression. EMBO J 2011, 30, 1289–1301. [Google Scholar]
- Gong, Z.; Brackertz, M.; Renkawitz, R. SUMO modification enhances p66-mediated transcriptional repression of the Mi-2/NuRD complex. Mol. Cell. Biol 2006, 26, 4519–4528. [Google Scholar]
- Rytinki, M.M.; Palvimo, J.J. SUMOylation modulates the transcription repressor function of RIP140. J. Biol. Chem 2008, 283, 11586–11595. [Google Scholar]
- Duverger, O.; Chen, S.X.; Lee, D.; Li, T.; Chock, P.B.; Morasso, M.I. SUMOylation of DLX3 by SUMO1 promotes its transcriptional activity. J. Cell. Biochem 2011, 112, 445–452. [Google Scholar]
- Chen, W.Y.; Lee, W.C.; Hsu, N.C.; Huang, F.; Chung, B.C. SUMO modification of repression domains modulates function of nuclear receptor 5A1 (steroidogenic factor-1). J. Biol. Chem 2004, 279, 38730–38735. [Google Scholar]
- Hammer, G.D.; Krylova, I.; Zhang, Y.; Darimont, B.D.; Simpson, K.; Weigel, N.L.; Ingraham, H.A. Phosphorylation of the nuclear receptor SF-1 modulates cofactor recruitment: Integration of hormone signaling in reproduction and stress. Mol. Cell 1999, 3, 521–526. [Google Scholar]
- Campbell, L.A.; Faivre, E.J.; Show, M.D.; Ingraham, J.G.; Flinders, J.; Gross, J.D.; Ingraham, H.A. Decreased recognition of SUMO-sensitive target genes following modification of SF-1 (NR5A1). Mol. Cell. Biol 2008, 28, 7476–7486. [Google Scholar]
- Yang, S.H.; Galanis, A.; Witty, J.; Sharrocks, A.D. An extended consensus motif enhances the specificity of substrate modification by SUMO. EMBO J 2006, 25, 5083–5093. [Google Scholar]
- Komatsu, T.; Mizusaki, H.; Mukai, T.; Ogawa, H.; Baba, D.; Shirakawa, M.; Hatakeyama, S.; Nakayama, K.I.; Yamamoto, H.; Kikuchi, A.; et al. Small ubiquitin-like modifier 1 (SUMO-1) modification of the synergy control motif of Ad4 binding protein/steroidogenic factor 1 (Ad4BP/SF-1) regulates synergistic transcription between Ad4BP/SF-1 and Sox9. Mol. Endocrinol 2004, 18, 2451–2462. [Google Scholar]
- Hsieh, Y.L.; Kuo, H.Y.; Chang, C.C.; Naik, M.T.; Liao, P.H.; Ho, C.C.; Huang, T.C.; Jeng, J.C.; Hsu, P.H.; Tsai, M.D.; et al. Ubc9 acetylation modulates distinct SUMO target modification and hypoxia response. EMBO J 2013, 32, 791–804. [Google Scholar]
- Matic, I.; Schimmel, J.; Hendriks, I.A.; van Santen, M.A.; van de Rijke, F.; van Dam, H.; Gnad, F.; Mann, M.; Vertegaal, A.C. Site-specific identification of SUMO-2 targets in cells reveals an inverted SUMOylation motif and a hydrophobic cluster SUMOylation motif. Mol. Cell 2010, 39, 641–652. [Google Scholar]
- Melchior, F. SUMO—Nonclassical ubiquitin. Annu. Rev. Cell. Dev. Biol 2000, 16, 591–626. [Google Scholar]
- Suda, N.; Shibata, H.; Kurihara, I.; Ikeda, Y.; Kobayashi, S.; Yokota, K.; Murai-Takeda, A.; Nakagawa, K.; Oya, M.; Murai, M.; et al. Coactivation of SF-1-mediated transcription of steroidogenic enzymes by Ubc9 and PIAS1. Endocrinology 2011, 152, 2266–2277. [Google Scholar]
- Sirianni, R.; Nogueira, E.; Bassett, M.H.; Carr, B.R.; Suzuki, T.; Pezzi, V.; Andò, S.; Rainey, W.E. The AP-1 family member FOS blocks transcriptional activity of the nuclear receptor steroidogenic factor 1. J. Cell. Sci 2010, 123, 3956–3965. [Google Scholar]
- Yang, W.H.; Gutierrez, N.M.; Wang, L.; Ellsworth, B.S.; Wang, C.M. Synergistic activation of the Mc2r promoter by FOXL2 and NR5A1 in mice. Biol. Reprod 2010, 83, 842–851. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Wang, C.-M.; Liu, R.; Wang, L.; Yang, W.-H. Acidic Residue Glu199 Increases SUMOylation Level of Nuclear Hormone Receptor NR5A1. Int. J. Mol. Sci. 2013, 14, 22331-22345. https://doi.org/10.3390/ijms141122331
Wang C-M, Liu R, Wang L, Yang W-H. Acidic Residue Glu199 Increases SUMOylation Level of Nuclear Hormone Receptor NR5A1. International Journal of Molecular Sciences. 2013; 14(11):22331-22345. https://doi.org/10.3390/ijms141122331
Chicago/Turabian StyleWang, Chiung-Min, Runhua Liu, Lizhong Wang, and Wei-Hsiung Yang. 2013. "Acidic Residue Glu199 Increases SUMOylation Level of Nuclear Hormone Receptor NR5A1" International Journal of Molecular Sciences 14, no. 11: 22331-22345. https://doi.org/10.3390/ijms141122331




