Multivalent Protein Assembly Using Monovalent Self-Assembling Building Blocks
Abstract
:1. Introduction
2. Results and Discussion
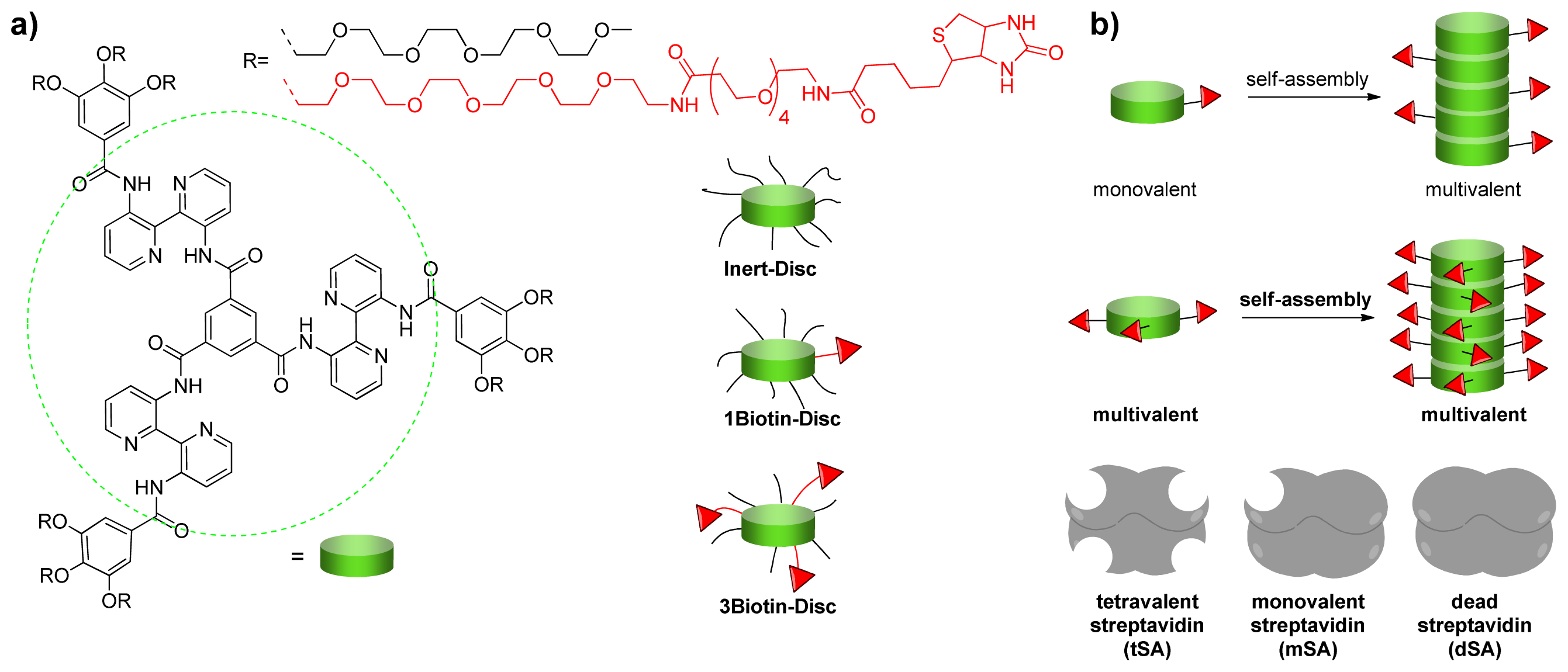
2.1. Design
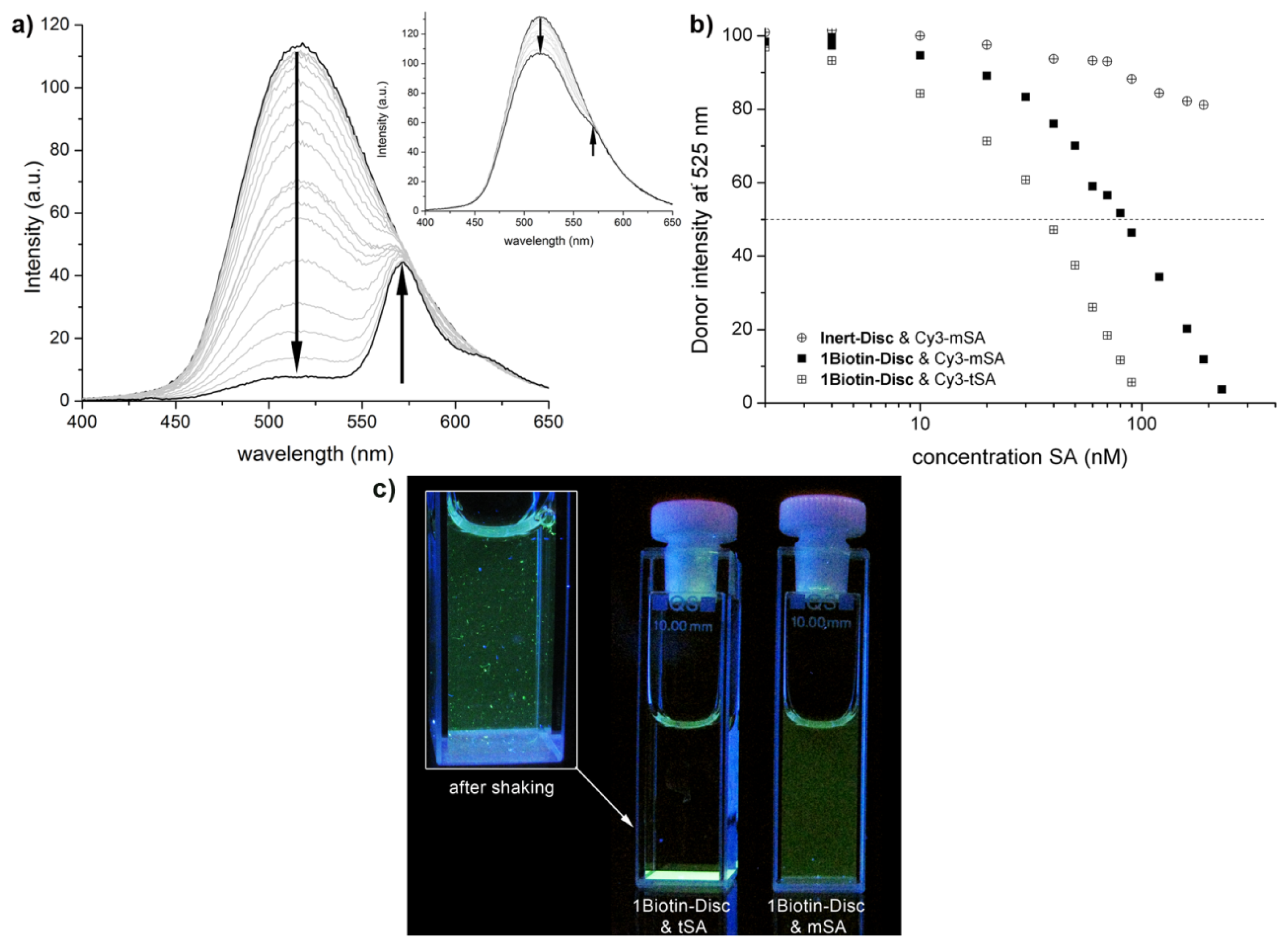
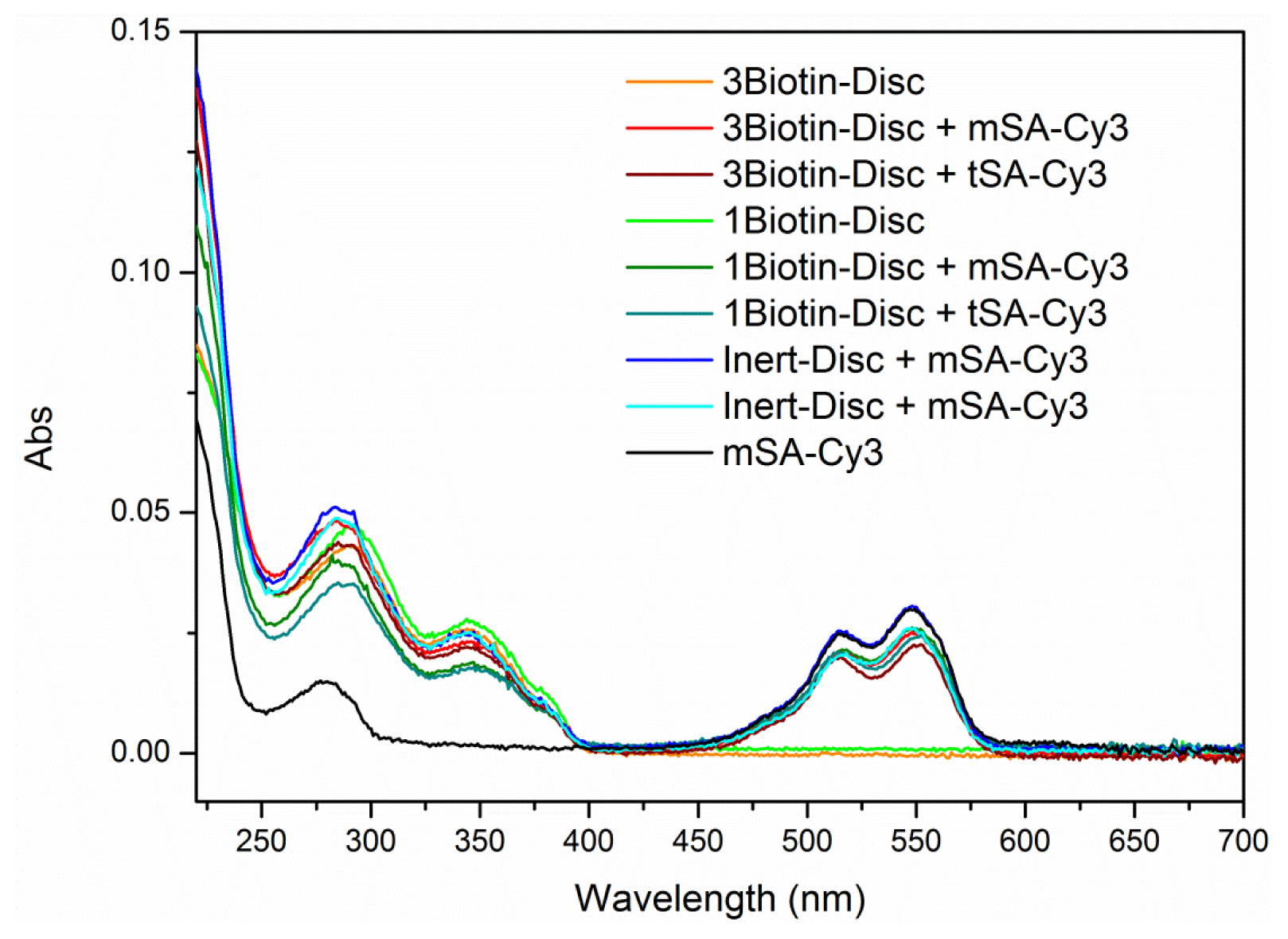
2.2. FRET Titrations
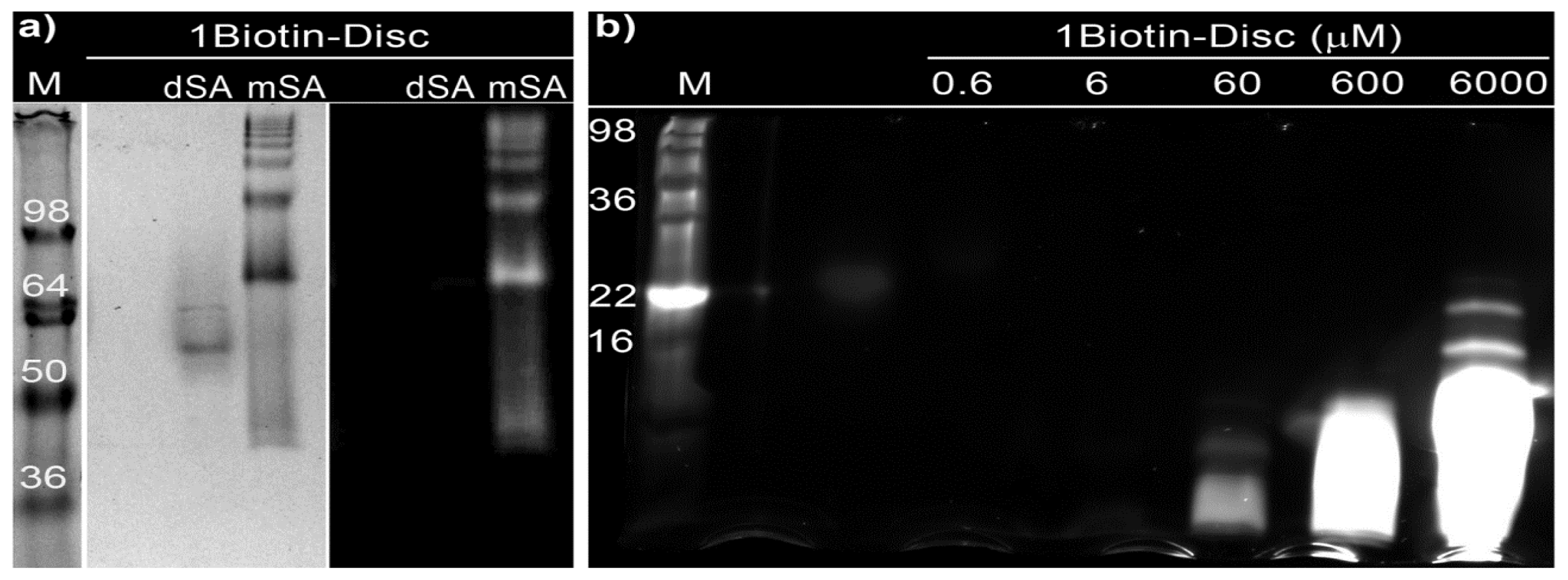
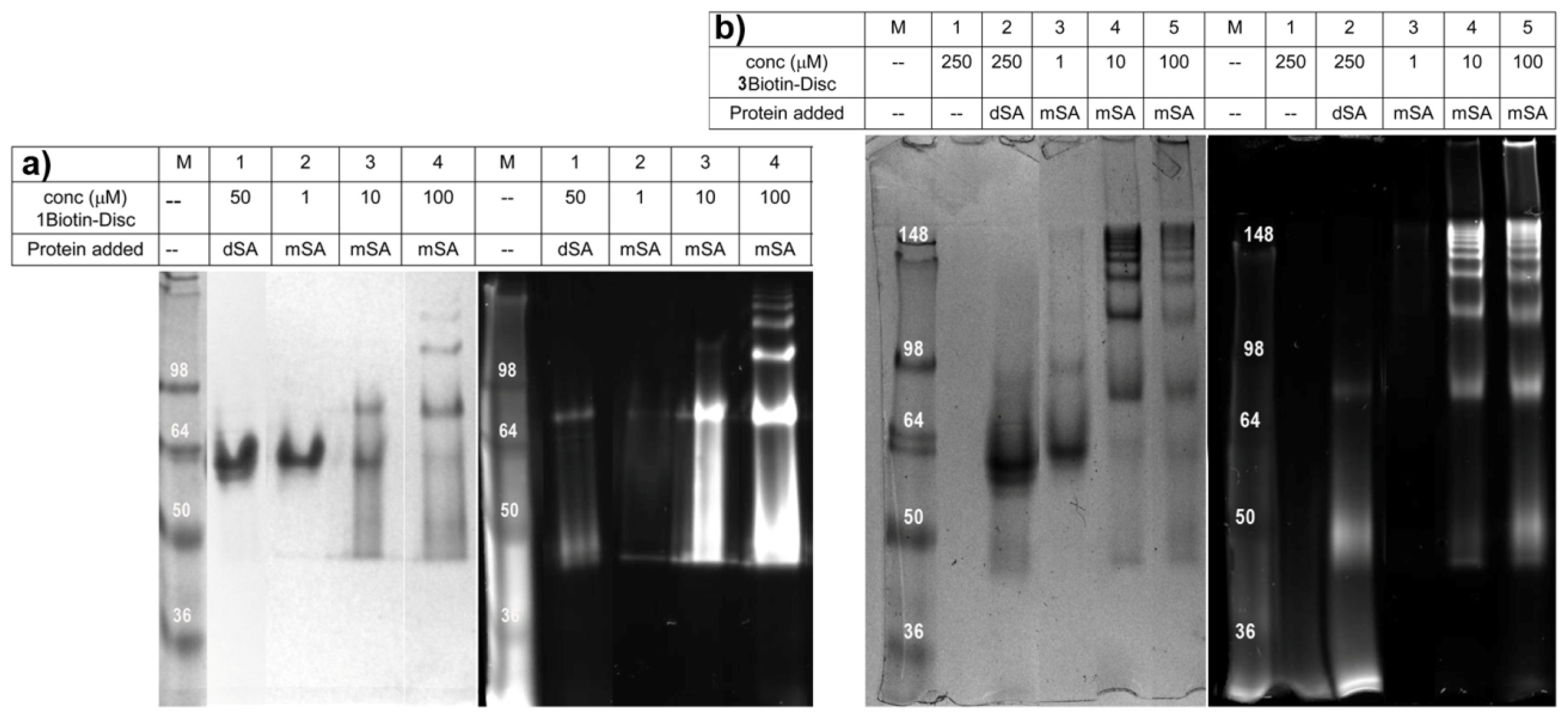
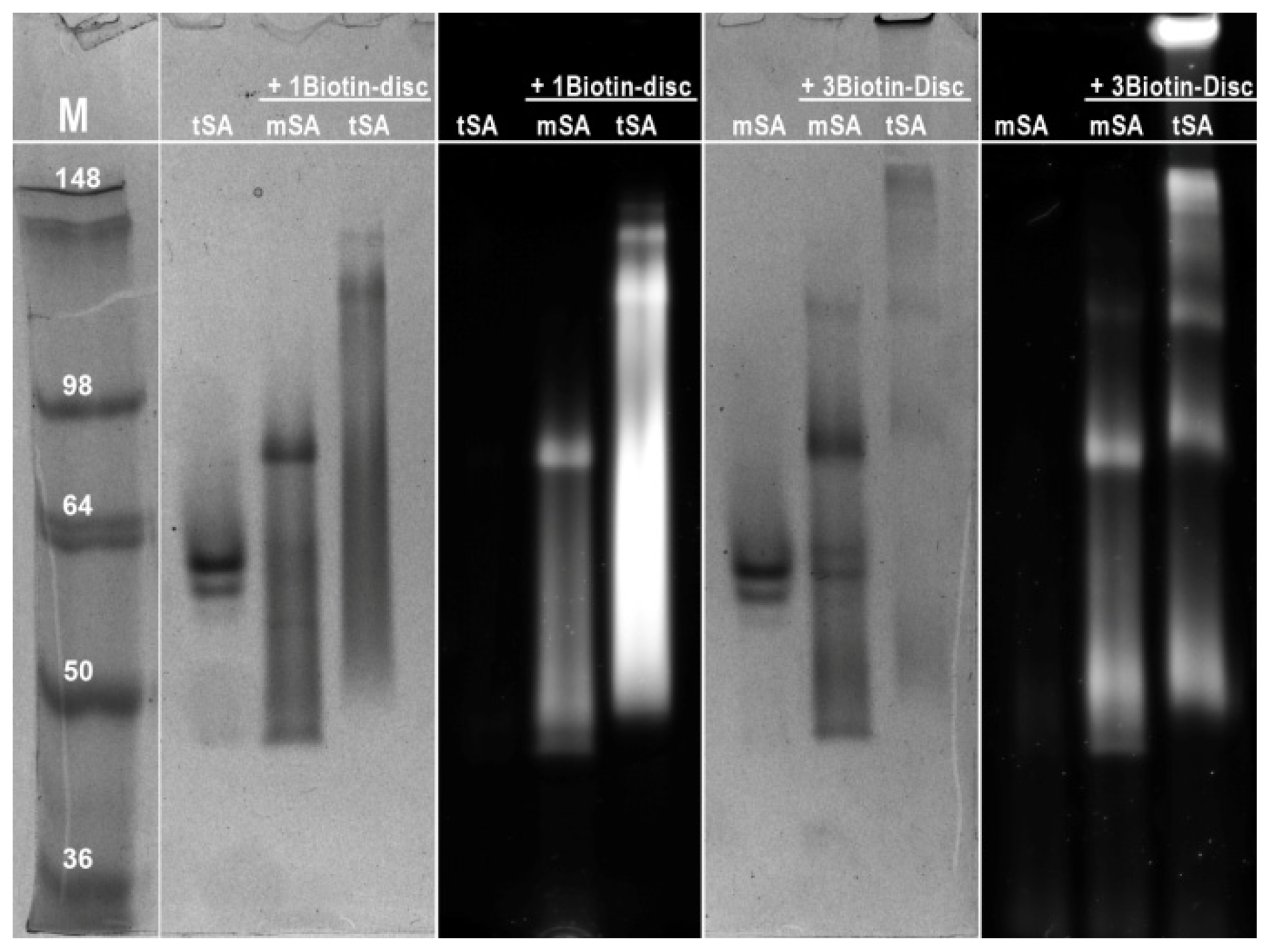
2.3. Semi-Native SDS-PAGE
3. Experimental Section
3.1. Materials and Methods
3.2. Synthesis of 1Biotin-Disc
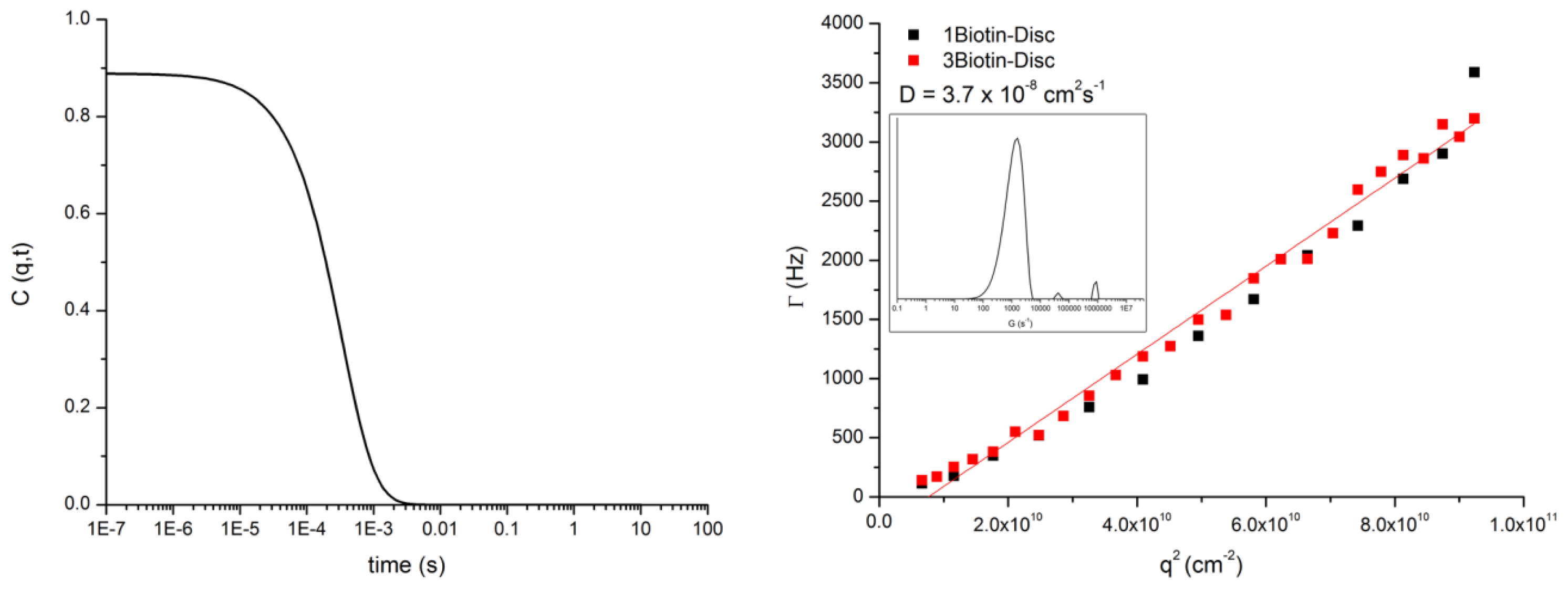
3.3. DLS of 1Biotin- and 3Biotin-Disc
3.4. Expression and Labeling of Streptavidin
3.5. FRET Titrations
3.6. Semi-Native SDS-PAGE
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Germain, R.N. T-cell signaling: The importance of receptor clustering. Curr. Biol 1997, 7, R640–R644. [Google Scholar]
- Pawson, T.; Nash, P. Assembly of cell regulatory systems through protein interaction domains. Science 2003, 300, 445–452. [Google Scholar]
- Taipale, J.; Keski-Oja, J. Growth factors in the extracellular matrix. FASEB J 1997, 11, 51–59. [Google Scholar]
- Pollard, T.D.; Cooper, J.A. Actin, a central player in cell shape and movement. Science 2009, 326, 1208–1212. [Google Scholar]
- Eisenberg, D.; Nelson, R.; Sawaya, M.R.; Balbirnie, M.; Sambashivan, S.; Ivanova, M.I.; Madsen, A.Ø.; Riekel, C. The structural biology of protein aggregation diseases: Fundamental questions and some answers. Acc. Chem. Res 2006, 39, 568–575. [Google Scholar]
- Levy, Y.; Onuchic, J.N. Mechanisms of protein assembly: Lessons from minimalist models. Acc. Chem. Res 2006, 39, 135–142. [Google Scholar]
- DeToma, A.S.; Salamekh, S.; Ramamoorthy, A.; Lim, M.H. Misfolded proteins in Alzheimer’s disease and type II diabetes. Chem. Soc. Rev 2012, 41, 608–621. [Google Scholar]
- Yeates, T.O.; Padilla, J.E. Designing supramolecular protein assemblies. Curr. Opin. Struct. Biol 2002, 12, 464–470. [Google Scholar]
- Salgado, E.N.; Radford, R.J.; Tezcan, F.A. Metal-directed protein self-assembly. Acc. Chem. Res 2010, 43, 661–672. [Google Scholar]
- Fegan, A.; White, B.; Carlson, J.C.T.; Wagner, C.R. Chemically controlled protein assembly: Techniques and applications. Chem. Rev 2010, 110, 3315–3336. [Google Scholar]
- Scheibel, T. Protein fibers as performance proteins: New technologies and applications. Curr. Opin. Biotech 2005, 16, 427–433. [Google Scholar]
- Patolsky, F.; Weizmann, Y.; Willner, I. Actin-based metallic nanowires as bio-nanotransporters. Nat. Mater 2004, 3, 692–695. [Google Scholar]
- Ringler, P.; Schulz, G.E. Self-assembly of proteins into designed networks. Science 2003, 302, 106–109. [Google Scholar]
- Dotan, N.; Arad, D.; Frolow, F.; Freeman, A. Self-assembly of a tetrahedral lectin into predesigned diamondlike protein crystals. Angew. Chem. Int. Ed 1999, 38, 2363–2366. [Google Scholar]
- Mori, Y.; Minamihata, K.; Abe, H.; Goto, M.; Kamiya, N. Protein assemblies by site-specific avidin–biotin interactions. Org. Biomol. Chem 2011, 9, 5641–5644. [Google Scholar]
- Ma, M.; Bong, D. Protein assembly directed by synthetic molecular recognition motifs. Org. Biomol. Chem 2011, 9, 7296–7299. [Google Scholar]
- Oohora, K.; Onoda, A.; Hayashi, T. Supramolecular assembling systems formed by heme–heme pocket interactions in hemoproteins. Chem. Commun 2012, 48, 11714–11726. [Google Scholar]
- Mori, Y.; Wakabayashi, R.; Goto, M.; Kamiya, N. Protein supramolecular complex formation by site-specific avidin–biotin interactions. Org. Biomol. Chem 2013, 11, 914–922. [Google Scholar]
- Burazerovic, S.; Gradinaru, J.; Pierron, J.; Ward, T.R. Hierarchical self-assembly of one-dimensional streptavidin bundles as a collagen mimetic for the biomineralization of calcite. Angew. Chem. Int. Ed 2007, 46, 5510–5514. [Google Scholar]
- Oohora, K.; Burazerovic, S.; Onoda, A.; Wilson, Y.M.; Ward, T.R.; Hayashi, T. Chemically programmed supramolecular assembly of hemoprotein and streptavidin with alternating alignment. Angew. Chem. Int. Ed 2012, 51, 3818–3821. [Google Scholar]
- Guler, M.O.; Soukasene, S.; Hulvat, J.F.; Stupp, S.I. Presentation and recognition of biotin on nanofibers formed by branched peptide amphiphiles. Nano Lett 2005, 5, 249–252. [Google Scholar]
- Müller, M.K.; Petkau, K.; Brunsveld, L. Protein assembly along a supramolecular wire. Chem. Commun 2011, 47, 310–312. [Google Scholar]
- Petkau-Milroy, K.; Uhlenheuer, D.A.; Spiering, A.J.H.; Vekemans, J.A.J.M.; Brunsveld, L. Dynamic and bio-orthogonal protein assembly along a supramolecular polymer. Chem. Sci 2013, 4, 2886–2891. [Google Scholar]
- Barnard, A.; Smith, D.K. Self-assembled multivalency: Dynamic ligand arrays for high-affinity binding. Angew. Chem. Int. Ed 2012, 51, 6572–6581. [Google Scholar]
- Thoma, G.; Katopodis, A.G.; Voelcker, N.; Duthaler, R.O.; Streiff, M.B. Novel glycodendrimers self-assemble to nanoparticles which function as polyvalent ligands in vitro and in vivo. Angew. Chem. Int. Ed. 2002, 41, 3195–3198. [Google Scholar]
- Toele, P.; van Gorp, J.J.; Glasbeek, M. Femtosecond fluorescence studies of self-assembled helical aggregates in solution. J. Phys. Chem. A 2005, 109, 10479–10487. [Google Scholar]
- Brunsveld, L.; Lohmeijer, B.G.G.; Vekemans, J.A.J.M.; Meijer, E.W. Chirality amplification in dynamic helical columns in water. Chem. Commun 2000, 2305–2306. [Google Scholar]
- Müller, M.K.; Brunsveld, L. A supramolecular polymer as a self-assembling polyvalent scaffold. Angew. Chem. Int. Ed 2009, 48, 2921–2924. [Google Scholar]
- Petkau-Milroy, K.; Brunsveld, L. Self-assembling multivalency–supramolecular polymers assembled from monovalent mannose-labelled discotic molecules. Eur. J. Org. Chem 2013, 2013, 3470–3476. [Google Scholar]
- Petkau-Milroy, K.; Sonntag, M.H.; van Onzen, A.H.A.M.; Brunsveld, L. Supramolecular polymers as dynamic multicomponent cellular uptake carriers. J. Am. Chem. Soc 2012, 134, 8086–8089. [Google Scholar]
- Green, N.M. Avidin and Streptavidin. In Avidin-Biotin Technology; Methods in Enzymology; Elsevier: Amsterdam, the Netherlands, 1990; Volume 184, pp. 51–67. [Google Scholar]
- Howarth, M.; Chinnapen, D.J.-F.; Gerrow, K.; Dorrestein, P.C.; Grandy, M.R.; Kelleher, N.L.; El-Husseini, A.; Ting, A.Y. A monovalent streptavidin with a single femtomolar biotin binding site. Nat. Meth 2006, 3, 267–273. [Google Scholar]
- Howarth, M.; Ting, A.Y. Monovalent streptavidin expression and purification. Nat. Protoc 2008. [Google Scholar] [CrossRef]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy; Springer: Heidelberg, Germany; p. 2006.
- Maurel, D.; Comps-Agrar, L.; Brock, C.; Rives, M.-L.; Bourrier, E.; Ayoub, M.A.; Bazin, H.; Tinel, N.; Durroux, T.; Prezeau, L.; et al. Cell-surface protein-protein interaction analysis with time-resolved FRET and snap-tag technologies: Application to GPCR oligomerization. Nat. Methods 2008, 5, 561–567. [Google Scholar]
- Huebsch, N.D.; Mooney, D.J. Fluorescent resonance energy transfer: A tool for probing molecular cell-biomaterial interactions in three dimensions. Biomaterials 2007, 28, 2424–2437. [Google Scholar]
- VanVeller, B.; Swager, T.M. Biocompatible post-polymerization functionalization of a water soluble poly(p-phenylene ethynylene). Chem. Commun 2010, 46, 5761–5763. [Google Scholar]
- Petkau-Milroy, K. Self-Assembling Auto-Fluorescent Amphiphiles; Technische Universiteit Eindhoven: Eindhoven, The Netherlands, 2012. [Google Scholar]
- Shapiro, A.L.; Viñuela, E.; Maizel, J.V., Jr. Molecular weight estimation of polypeptide chains by electrophoresis in SDS-polyacrylamide gels. Biochem. Biophys. Res. Commun 1967, 28, 815–820. [Google Scholar]
- Le Gac, S.; Schwartz, E.; Koepf, M.; Cornelissen, J.J.L.M.; Rowan, A.E.; Nolte, R.J.M. Cysteine-Containing polyisocyanides as versatile nanoplatforms for chromophoric and bioscaffolding. Chem. Eur. J 2010, 16, 6176–6186. [Google Scholar]
- Brunsveld, L.; Folmer, B.J.B.; Meijer, E.W.; Sijbesma, R.P. Supramolecular polymers. Chem. Rev 2001, 101, 4071–4098. [Google Scholar]
- Melville,, D.B. Biotin Sulfoxide. J. Biol. Chem 1954, 208, 495–502. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Petkau-Milroy, K.; Sonntag, M.H.; Colditz, A.; Brunsveld, L. Multivalent Protein Assembly Using Monovalent Self-Assembling Building Blocks. Int. J. Mol. Sci. 2013, 14, 21189-21201. https://doi.org/10.3390/ijms141021189
Petkau-Milroy K, Sonntag MH, Colditz A, Brunsveld L. Multivalent Protein Assembly Using Monovalent Self-Assembling Building Blocks. International Journal of Molecular Sciences. 2013; 14(10):21189-21201. https://doi.org/10.3390/ijms141021189
Chicago/Turabian StylePetkau-Milroy, Katja, Michael H. Sonntag, Alexander Colditz, and Luc Brunsveld. 2013. "Multivalent Protein Assembly Using Monovalent Self-Assembling Building Blocks" International Journal of Molecular Sciences 14, no. 10: 21189-21201. https://doi.org/10.3390/ijms141021189



