Biofilm Matrix and Its Regulation in Pseudomonas aeruginosa
Abstract
:1. Introduction
2. Matrices of P. aeruginosa Biofilms
2.1. Psl Polysaccharide
2.2. Pel Polysaccharide
2.3. Alginate
2.4. Extracellular DNA
2.5. Proteins and Proteinaceous Bacterial Surface Appendages
3. Regulation of Biofilm Matrix in P. aeruginosa
3.1. c-di-GMP
3.2. GacA/GacS Two-Component Systems
3.3. Quorum Sensing
3.4. Other Types of Regulation
4. Matrix-Driven Strategies against Biofilms
5. Perspectives

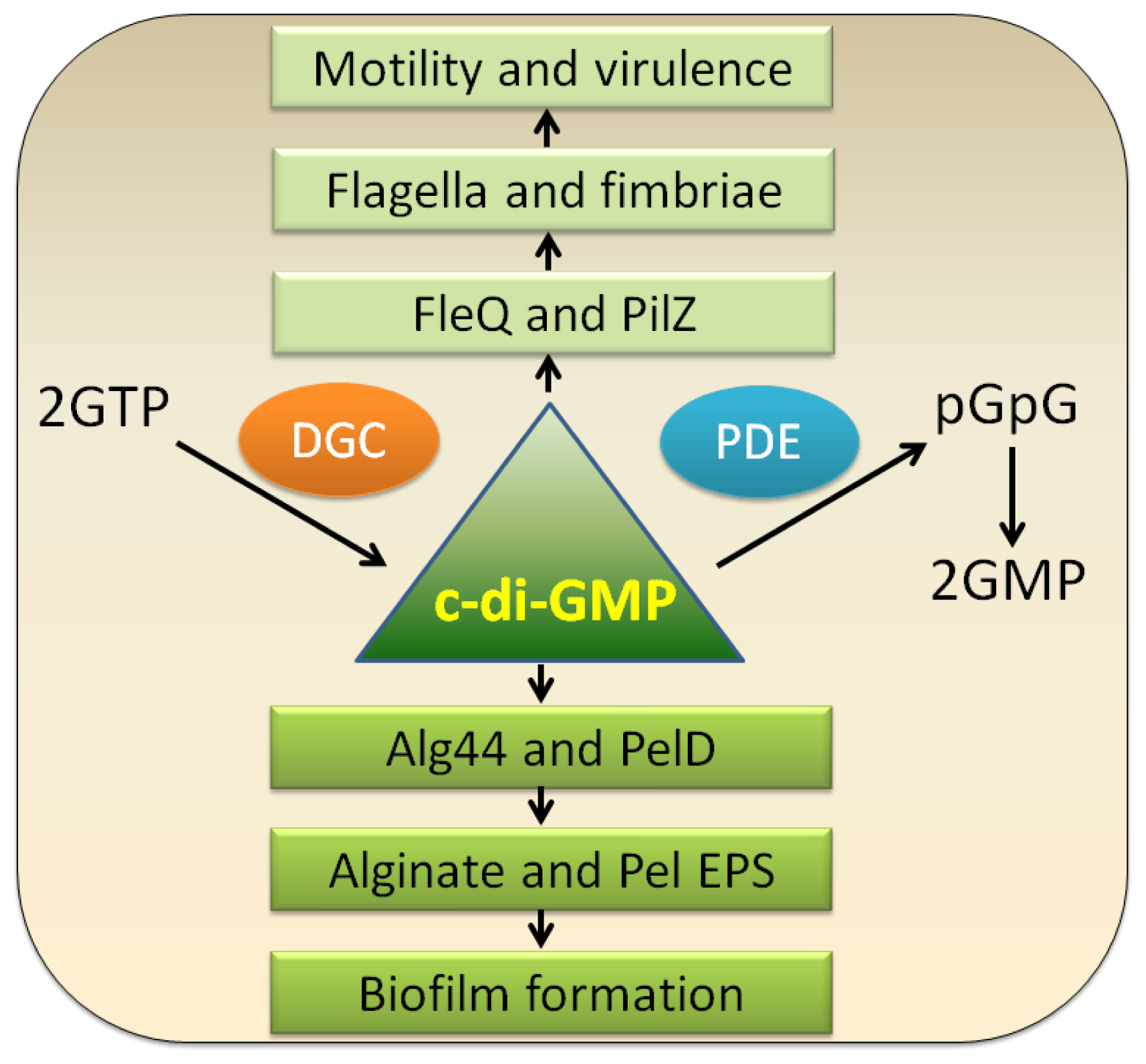
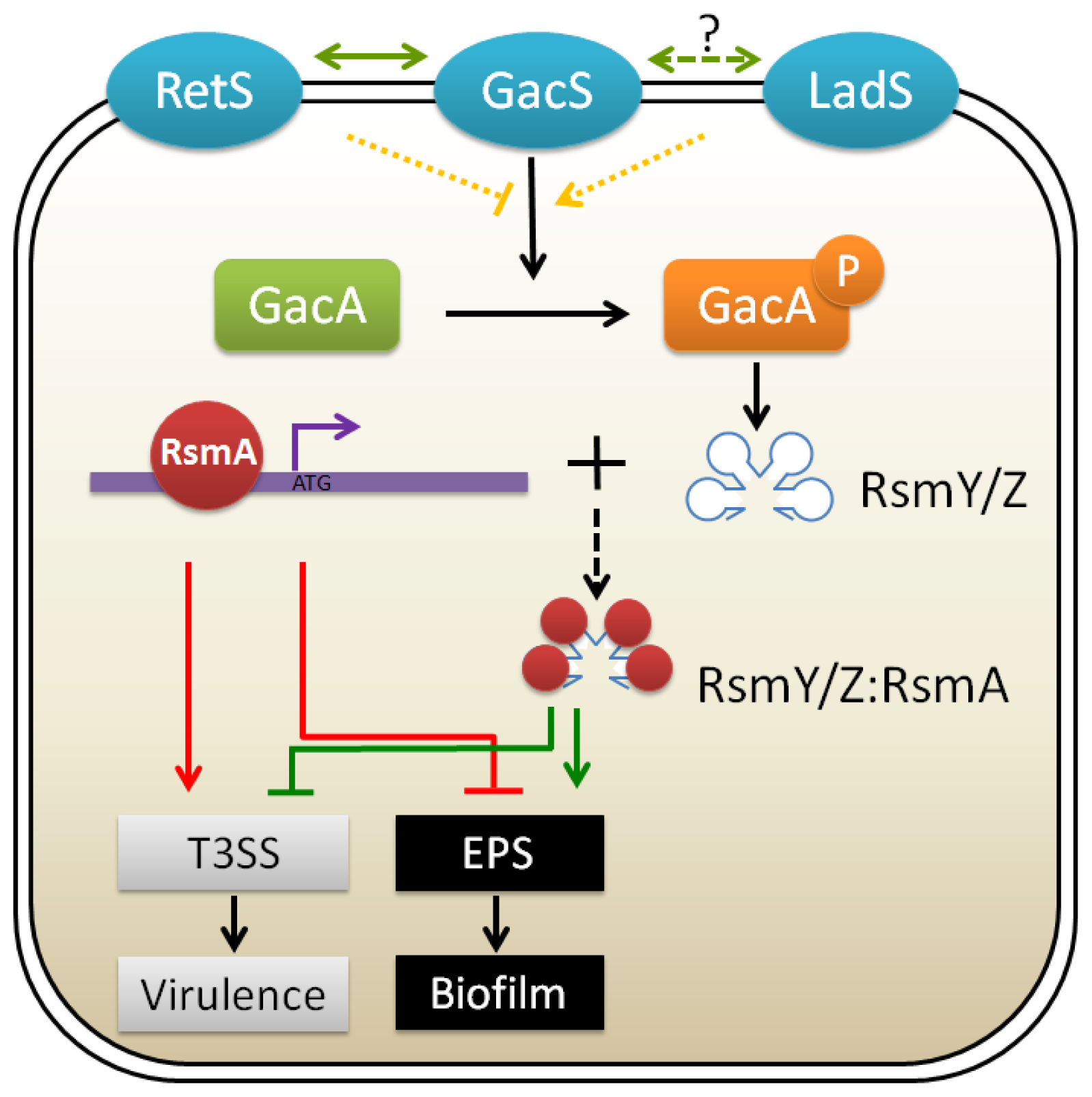
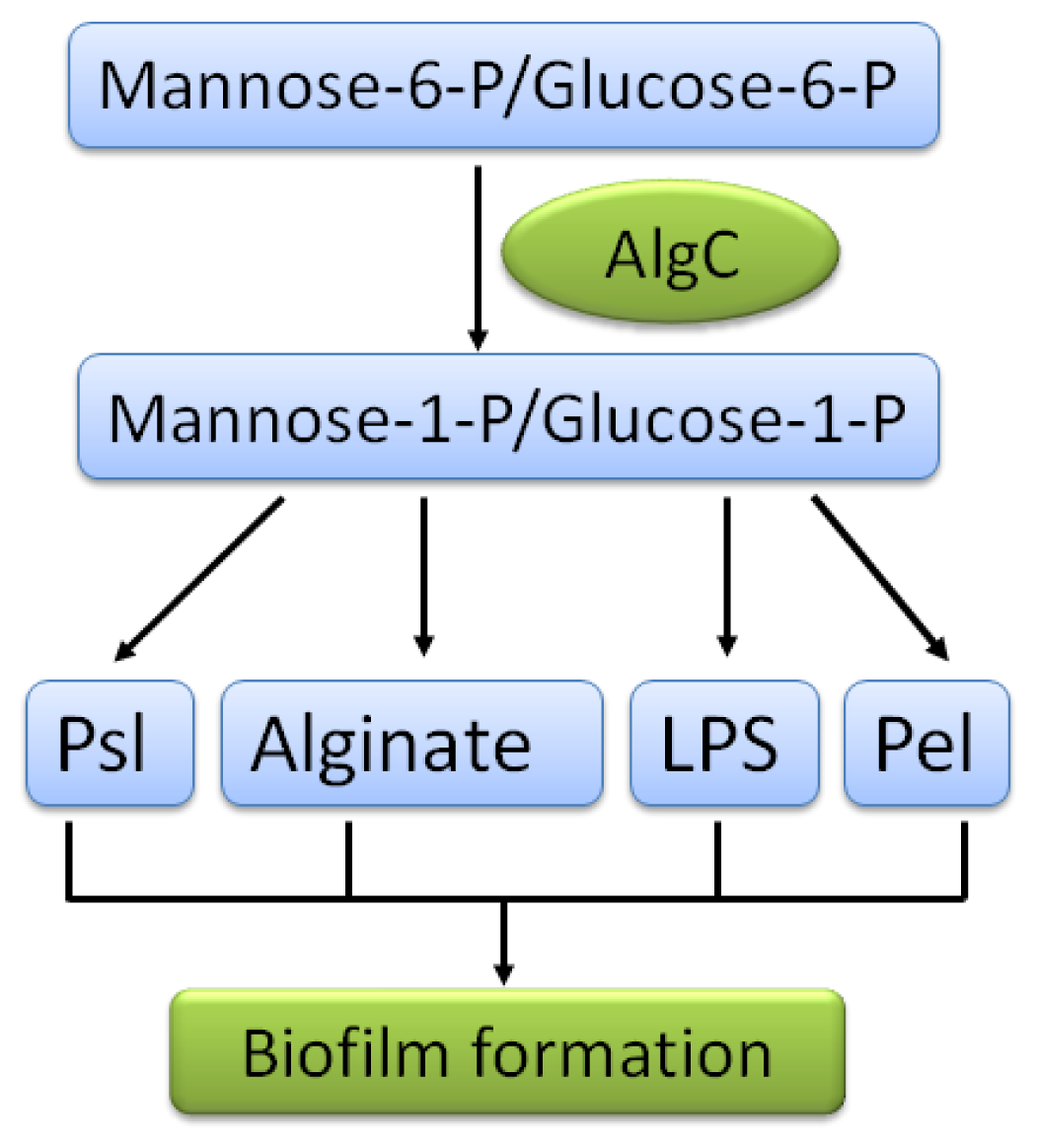
| EPS | Locus * | Roles | References |
|---|---|---|---|
| Psl | PA2231–PA2245 | Initial attachment and adhesion | [15,17–19] |
| Primary biofilm scaffold | [15,19] | ||
| Proinflammatory signaling | [20] | ||
| Antibiotics resistance | [19] | ||
| Avoidance of host defence mechanisms | [30] | ||
| Signaling molecule to stimulate biofilm formation | [28] | ||
| Resistance to biofilm inhibitor Polysorbate 80 | [31] | ||
| Guide of exploration and microcolony formation | [26] | ||
| Pel | PA3058–PA3064 | Pellicle formation and solid surface-associated biofilm formation | [11] |
| Aggregating of bacterial cells | [14] | ||
| Aminoglycosides antibiotic resistance | [14] | ||
| Initial attachment in the absence of type IV pili | [34] | ||
| Alginate | PA3540–PA3548 | Persistence and immune evasion | [44] |
| Resistance to antibiotics as well as opsonophagocytosis | [45,46] | ||
| ROS scavenge | [47] | ||
| Leading to mucoid | [39] | ||
| Water and nutrient retention | [41] | ||
| eDNA | Nutrient | [54,55] | |
| Scaffold | [49] | ||
| Antibiotics resistance | [53] | ||
| Major proinflammatory component | [57] | ||
| Promoting self-organization of bacterial biofilms | [58] | ||
| Proteinaceous component | Flagella | Initial cell-to-surface interactions | [60] |
| Pili | The formation of mushroom-like microcolony | [60] | |
| CdrA | Mediate aggregation and increase biofilm stability | [63] | |
| Cup fimbriae | Cell-to-cell interaction and microcolony formation | [66,67] | |
Acknowledgments
Conflicts of Interest
References
- Karatan, E.; Watnick, P. Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol. Mol. Biol Rev 2009, 73, 310–347. [Google Scholar]
- Davies, D.G.; Parsek, M.R.; Pearson, J.P.; Iglewski, B.H.; Costerton, J.W.; Greenberg, E.P. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 1998, 280, 295–298. [Google Scholar]
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial biofilms: From the natural environment to infectious diseases. Nat. Rev 2004, 2, 95–108. [Google Scholar]
- Sutherland, I.W. The biofilm matrix—An immobilized but dynamic microbial environment. Trends Microbiol 2001, 9, 222–227. [Google Scholar]
- Branda, S.S.; Vik, S.; Friedman, L.; Kolter, R. Biofilms: The matrix revisited. Trends Microbiol 2005, 13, 20–26. [Google Scholar]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial biofilms: A common cause of persistent infections. Science 1999, 284, 1318–1322. [Google Scholar]
- Drenkard, E.; Ausubel, F.M. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature 2002, 416, 740–743. [Google Scholar]
- Flemming, H.C.; Neu, T.R.; Wozniak, D.J. The EPS matrix: The “house of biofilm cells”. J. Bacteriol 2007, 189, 7945–7947. [Google Scholar]
- Ryder, C.; Byrd, M.; Wozniak, D.J. Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr. Opin. Microbiol 2007, 10, 644–648. [Google Scholar]
- Parsek, M.R.; Singh, P.K. Bacterial biofilms: An emerging link to disease pathogenesis. Annu. Rev. Microbiol 2003, 57, 677–701. [Google Scholar]
- Friedman, L.; Kolter, R. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J. Bacteriol 2004, 186, 4457–4465. [Google Scholar]
- Friedman, L.; Kolter, R. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol 2004, 51, 675–690. [Google Scholar]
- Jackson, K.D.; Starkey, M.; Kremer, S.; Parsek, M.R.; Wozniak, D.J. Identification of psl, a locus encoding a potential exopolysaccharide that is essential for Pseudomonas aeruginosa PAO1 biofilm formation. J. Bacteriol 2004, 186, 4466–4475. [Google Scholar]
- Colvin, K.M.; Gordon, V.D.; Murakami, K.; Borlee, B.R.; Wozniak, D.J.; Wong, G.C.; Parsek, M.R. The pel polysaccharide can serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa. PLoS Pathog 2010, 7, e1001264. [Google Scholar]
- Ma, L.; Conover, M.; Lu, H.; Parsek, M.R.; Bayles, K.; Wozniak, D.J. Assembly and development of the Pseudomonas aeruginosa biofilm matrix. PLoS Pathog 2009, 5, e1000354. [Google Scholar]
- Matsukawa, M.; Greenberg, E.P. Putative exopolysaccharide synthesis genes influence Pseudomonas aeruginosa biofilm development. J. Bacteriol 2004, 186, 4449–4456. [Google Scholar]
- Ma, L.; Lu, H.; Sprinkle, A.; Parsek, M.R.; Wozniak, D.J. Pseudomonas aeruginosa Psl is a galactose- and mannose-rich exopolysaccharide. J. Bacteriol 2007, 189, 8353–8356. [Google Scholar]
- Ma, L.; Jackson, K.D.; Landry, R.M.; Parsek, M.R.; Wozniak, D.J. Analysis of Pseudomonas aeruginosa conditional psl variants reveals roles for the psl polysaccharide in adhesion and maintaining biofilm structure postattachment. J. Bacteriol 2006, 188, 8213–8221. [Google Scholar]
- Yang, L.; Hu, Y.F.; Liu, Y.; Zhang, J.D.; Ulstrup, J.; Molin, S. Distinct roles of extracellular polymeric substances in Pseudomonas aeruginosa biofilm development. Environ. Microbiol 2011, 13, 1705–1717. [Google Scholar]
- Byrd, M.S.; Sadovskaya, I.; Vinogradov, E.; Lu, H.P.; Sprinkle, A.B.; Richardson, S.H.; Ma, L.Y.; Ralston, B.; Parsek, M.R.; Anderson, E.M.; et al. Genetic and biochemical analyses of the Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in Psl and LPS production. Mol. Microbiol 2009, 73, 622–638. [Google Scholar]
- Winsor, G.L.; Lam, D.K.; Fleming, L.; Lo, R.; Whiteside, M.D.; Yu, N.Y.; Hancock, R.E.; Brinkman, F.S. Pseudomonas Genome Database: Improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res 2011, 39, D596–D600. [Google Scholar]
- Campisano, A.; Schroeder, C.; Schemionek, M.; Overhage, J.; Rehm, B.H. PslD is a secreted protein required for biofilm formation by Pseudomonas aeruginosa. Appl. Environ. Microbiol 2006, 72, 3066–3068. [Google Scholar]
- Overhage, J.; Schemionek, M.; Webb, J.S.; Rehm, B.H. Expression of the psl operon in Pseudomonas aeruginosa PAO1 biofilms: PslA performs an essential function in biofilm formation. Appl. Environ. Microbiol 2005, 71, 4407–4413. [Google Scholar]
- Lee, H.J.; Chang, H.Y.; Venkatesan, N.; Peng, H.L. Identification of amino acid residues important for the phosphomannose isomerase activity of PslB in Pseudomonas aeruginosa PAO1. FEBS Lett 2008, 582, 3479–3483. [Google Scholar]
- Wang, S.; Parsek, M.R.; Wozniak, D.J.; Ma, L.Z. A spider web strategy of type IV pili-mediated migration to build a fibre-like Psl polysaccharide matrix in Pseudomonas aeruginosa biofilms. Environ. Microbiol 2013, 15, 2238–2253. [Google Scholar]
- Zhao, K.; Tseng, B.S.; Beckerman, B.; Jin, F.; Gibiansky, M.L.; Harrison, J.J.; Luijten, E.; Parsek, M.R.; Wong, G.C. Psl trails guide exploration and microcolony formation in Pseudomonas aeruginosa biofilms. Nature 2013, 497, 388–391. [Google Scholar]
- Stoodley, P.; Sauer, K.; Davies, D.G.; Costerton, J.W. Biofilms as complex differentiated communities. Annu. Rev. Microbiol 2002, 56, 187–209. [Google Scholar]
- Irie, Y.; Borlee, B.R.; O’Connor, J.R.; Hill, P.J.; Harwood, C.S.; Wozniak, D.J.; Parsek, M.R. Self-produced exopolysaccharide is a signal that stimulates biofilm formation in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2012, 109, 20632–20636. [Google Scholar]
- Byrd, M.S.; Pang, B.; Mishra, M.; Swords, W.E.; Wozniak, D.J. The Pseudomonas aeruginosa exopolysaccharide Psl facilitates surface adherence and NF-kappaB activation in A549 cells. mBiosphere 2010, 1. [Google Scholar] [CrossRef]
- Mishra, M.; Byrd, M.S.; Sergeant, S.; Azad, A.K.; Parsek, M.R.; McPhail, L.; Schlesinger, L.S.; Wozniak, D.J. Pseudomonas aeruginosa Psl polysaccharide reduces neutrophil phagocytosis and the oxidative response by limiting complement-mediated opsonization. Cell Microbiol 2012, 14, 95–106. [Google Scholar]
- Zegans, M.E.; Wozniak, D.; Griffin, E.; Toutain-Kidd, C.M.; Hammond, J.H.; Garfoot, A.; Lam, J.S. Pseudomonas aeruginosa exopolysaccharide Psl promotes resistance to the biofilm inhibitor polysorbate 80. Antimicrob. Agents Chemother. 2012, 56, 4112–4122. [Google Scholar]
- Tseng, B.S.; Zhang, W.; Harrison, J.J.; Quach, T.P.; Song, J.L.; Penterman, J.; Singh, P.K.; Chopp, D.L.; Packman, A.I.; Parsek, M.R. The extracellular matrix protects Pseudomonas aeruginosa biofilms by limiting the penetration of tobramycin. Environ. Microbiol 2013, 15, 2865–2878. [Google Scholar]
- Billings, N.; Ramirez Millan, M.; Caldara, M.; Rusconi, R.; Tarasova, Y.; Stocker, R.; Ribbeck, K. The extracellular matrix component Psl provides fast-acting antibiotic defense in Pseudomonas aeruginosa biofilms. PLoS Pathog 2013, 9, e1003526. [Google Scholar]
- Vasseur, P.; Vallet-Gely, I.; Soscia, C.; Genin, S.; Filloux, A. The pel genes of the Pseudomonas aeruginosa PAK strain are involved at early and late stages of biofilm formation. Microbiology 2005, 151, 985–997. [Google Scholar]
- Sadovskaya, I.; Vinogradov, E.; Li, J.J.; Hachani, A.; Kowalska, K.; Filloux, A. High-level antibiotic resistance in Pseudomonas aeruginosa biofilm: The ndvB gene is involved in the production of highly glycerol-phosphorylated beta-(1->3)-glucans, which bind aminoglycosides. Glycobiology 2010, 20, 895–904. [Google Scholar]
- Coulon, C.; Vinogradov, E.; Filloux, A.; Sadovskaya, I. Chemical analysis of cellular and extracellular carbohydrates of a biofilm-forming strain Pseudomonas aeruginosa PA14. PLoS One 2010, 5, e14220. [Google Scholar]
- Ghafoor, A.; Jordens, Z.; Rehm, B.H. Role of PelF in Pel polysaccharide biosynthesis in Pseudomonas aeruginosa. Appl. Environ. Microbiol 2013, 79, 2968–2978. [Google Scholar]
- Colvin, K.M.; Alnabelseya, N.; Baker, P.; Whitney, J.C.; Howell, P.L.; Parsek, M.R. PelA deacetylase activity is required for Pel polysaccharide synthesis in Pseudomonas aeruginosa. J. Bacteriol 2013, 195, 2329–2339. [Google Scholar]
- Govan, J.R.; Deretic, V. Microbial pathogenesis in cystic fibrosis: Mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev 1996, 60, 539–574. [Google Scholar]
- Wozniak, D.J.; Wyckoff, T.J.; Starkey, M.; Keyser, R.; Azadi, P.; O’Toole, G.A.; Parsek, M.R. Alginate is not a significant component of the extracellular polysaccharide matrix of PA14 and PAO1 Pseudomonas aeruginosa biofilms. Proc. Natl. Acad. Sci. USA 2003, 100, 7907–7912. [Google Scholar]
- Sutherland, I.W. Biofilm exopolysaccharides: A strong and sticky framework. Microbiology 2001, 147, 3–9. [Google Scholar]
- Ma, L.; Wang, S.; Wang, D.; Parsek, M.R.; Wozniak, D.J. The roles of biofilm matrix polysaccharide Psl in mucoid Pseudomonas aeruginosa biofilms. FEMS Immunol. Med. Microbiol 2012, 65, 377–380. [Google Scholar]
- Yang, L.; Hengzhuang, W.; Wu, H.; Damkiaer, S.; Jochumsen, N.; Song, Z.; Givskov, M.; Hoiby, N.; Molin, S. Polysaccharides serve as scaffold of biofilms formed by mucoid Pseudomonas aeruginosa. FEMS Immunol. Med. Microbiol 2012, 65, 366–376. [Google Scholar]
- Leid, J.G.; Willson, C.J.; Shirtliff, M.E.; Hassett, D.J.; Parsek, M.R.; Jeffers, A.K. The exopolysaccharide alginate protects Pseudomonas aeruginosa biofilm bacteria from IFN-gamma-mediated macrophage killing. J. Immunol 2005, 175, 7512–7518. [Google Scholar]
- Simpson, J.A.; Smith, S.E.; Dean, R.T. Alginate inhibition of the uptake of Pseudomonas aeruginosa by macrophages. J. Gen. Microbiol 1988, 134, 29–36. [Google Scholar]
- Simpson, J.A.; Smith, S.E.; Dean, R.T. Alginate may accumulate in cystic-fibrosis lung because the enzymatic and free-radical capacities of phagocytic-cells are inadequate for its degradation. Biochem. Mol. Biol Int 1993, 30, 1021–1034. [Google Scholar]
- Simpson, J.A.; Smith, S.E.; Dean, R.T. Scavenging by alginate of free-radicals released by macrophages. Free Radic. Bio. Med 1989, 6, 347–353. [Google Scholar]
- Bragonzi, A.; Paroni, M.; Nonis, A.; Cramer, N.; Montanari, S.; Rejman, J.; Di Serio, C.; Doring, G.; Tummler, B. Pseudomonas aeruginosa microevolution during cystic fibrosis lung infection establishes clones with adapted virulence. Am. J. Resp. Crit. Care 2009, 180, 138–145. [Google Scholar]
- Allesen-Holm, M.; Barken, K.B.; Yang, L.; Klausen, M.; Webb, J.S.; Kjelleberg, S.; Molin, S.; Givskov, M.; Tolker-Nielsen, T. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol. Microbiol 2006, 59, 1114–1128. [Google Scholar]
- Whitchurch, C.B.; Tolker-Nielsen, T.; Ragas, P.C.; Mattick, J.S. Extracellular DNA required for bacterial biofilm formation. Science 2002, 295, 1487. [Google Scholar]
- Webb, J.S.; Thompson, L.S.; James, S.; Charlton, T.; Tolker-Nielsen, T.; Koch, B.; Givskov, M.; Kjelleberg, S. Cell death in Pseudomonas aeruginosa biofilm development. J. Bacteriol 2003, 185, 4585–4592. [Google Scholar]
- Spoering, A.L.; Gilmore, M.S. Quorum sensing and DNA release in bacterial biofilms. Curr. Opin. Microbiol 2006, 9, 133–137. [Google Scholar]
- Mulcahy, H.; Charron-Mazenod, L.; Lewenza, S. Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog 2008, 4, e1000213. [Google Scholar]
- Finkel, S.E.; Kolter, R. DNA as a nutrient: Novel role for bacterial competence gene homologs. J. Bacteriol 2001, 183, 6288–6293. [Google Scholar]
- Mulcahy, H.; Charron-Mazenod, L.; Lewenza, S. Pseudomonas aeruginosa produces an extracellular deoxyribonuclease that is required for utilization of DNA as a nutrient source. Environ. Microbiol 2010, 12, 1621–1629. [Google Scholar]
- Parks, Q.M.; Young, R.L.; Poch, K.R.; Malcolm, K.C.; Vasil, M.L.; Nick, J.A. Neutrophil enhancement of Pseudomonas aeruginosa biofilm development: Human F-actin and DNA as targets for therapy. J. Med. Microbiol 2009, 58, 492–502. [Google Scholar]
- Fuxman Bass, J.I.; Russo, D.M.; Gabelloni, M.L.; Geffner, J.R.; Giordano, M.; Catalano, M.; Zorreguieta, A.; Trevani, A.S.; Extracellular, DNA. A major proinflammatory component of Pseudomonas aeruginosa biofilms. J. Immunol 2010, 184, 6386–6395. [Google Scholar]
- Gloag, E.S.; Turnbull, L.; Huang, A.; Vallotton, P.; Wang, H.; Nolan, L.M.; Mililli, L.; Hunt, C.; Lu, J.; Osvath, S.R.; et al. Self-organization of bacterial biofilms is facilitated by extracellular DNA. Proc. Natl. Acad. Sci. USA 2013, 110, 11541–11546. [Google Scholar]
- Mann, E.E.; Wozniak, D.J. Pseudomonas biofilm matrix composition and niche biology. Fems. Microbiol. Rev 2012, 36, 893–916. [Google Scholar]
- O’Toole, G.A.; Kolter, R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol 1998, 30, 295–304. [Google Scholar]
- Skerker, J.M.; Berg, H.C. Direct observation of extension and retraction of type IV pili. Proc. Natl. Acad. Sci. USA 2001, 98, 6901–6904. [Google Scholar]
- Klausen, M.; Heydorn, A.; Ragas, P.; Lambertsen, L.; Aaes-Jorgensen, A.; Molin, S.; Tolker-Nielsen, T. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol. Microbiol 2003, 48, 1511–1524. [Google Scholar]
- Borlee, B.R.; Goldman, A.D.; Murakami, K.; Samudrala, R.; Wozniak, D.J.; Parsek, M.R. Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol. Microbiol 2010, 75, 827–842. [Google Scholar]
- Vallet, I.; Olson, J.W.; Lory, S.; Lazdunski, A.; Filloux, A. The chaperone/usher pathways of Pseudomonas aeruginosa: Identification of fimbrial gene clusters (cup) and their involvement in biofilm formation. Proc. Natl. Acad. Sci. USA 2001, 98, 6911–6916. [Google Scholar]
- Giraud, C.; Bernard, C.S.; Calderon, V.; Yang, L.; Filloux, A.; Molin, S.; Fichant, G.; Bordi, C.; De Bentzmann, S. The PprA-PprB two-component system activates CupE, the first non-archetypal Pseudomonas aeruginosa chaperone-usher pathway system assembling fimbriae. Environ. Microbiol 2011, 13, 666–683. [Google Scholar]
- Kulasekara, H.D.; Ventre, I.; Kulasekara, B.R.; Lazdunski, A.; Filloux, A.; Lory, S. A novel two-component system controls the expression of Pseudomonas aeruginosa fimbrial cup genes. Mol. Microbiol 2005, 55, 368–380. [Google Scholar]
- Ruer, S.; Stender, S.; Filloux, A.; De Bentzmann, S. Assembly of fimbrial structures in Pseudomonas aeruginosa: Functionality and specificity of chaperone-usher machineries. J. Bacteriol 2007, 189, 3547–3555. [Google Scholar]
- Hoffman, L.R.; D’Argenio, D.A.; MacCoss, M.J.; Zhang, Z.; Jones, R.A.; Miller, S.I. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 2005, 436, 1171–1175. [Google Scholar]
- Whiteley, M.; Bangera, M.G.; Bumgarner, R.E.; Parsek, M.R.; Teitzel, G.M.; Lory, S.; Greenberg, E.P. Gene expression in Pseudomonas aeruginosa biofilms. Nature 2001, 413, 860–864. [Google Scholar]
- Ross, P.; Weinhouse, H.; Aloni, Y.; Michaeli, D.; Weinberger-Ohana, P.; Mayer, R.; Braun, S.; De Vroom, E.; Van der Marel, G.A.; Van Boom, J.H.; et al. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 1987, 325, 279–281. [Google Scholar]
- Hengge, R. Principles of c-di-GMP signalling in bacteria. Nat. Rev 2009, 7, 263–273. [Google Scholar]
- Cotter, P.A.; Stibitz, S. c-di-GMP-mediated regulation of virulence and biofilm formation. Curr. Opin. Microbiol 2007, 10, 17–23. [Google Scholar]
- Jenal, U.; Malone, J. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu. Rev. Genet 2006, 40, 385–407. [Google Scholar]
- Merighi, M.; Lee, V.T.; Hyodo, M.; Hayakawa, Y.; Lory, S. The second messenger bis-(3′-5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol. Microbiol 2007, 65, 876–895. [Google Scholar]
- Romling, U.; Galperin, M.Y.; Gomelsky, M. Cyclic di-GMP: The first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol Rev 2013, 77, 1–52. [Google Scholar]
- Ryan, R.P.; Lucey, J.; O’Donovan, K.; McCarthy, Y.; Yang, L.; Tolker-Nielsen, T.; Dow, J.M. HD-GYP domain proteins regulate biofilm formation and virulence in Pseudomonas aeruginosa. Environ. Microbiol 2009, 11, 1126–1136. [Google Scholar]
- Kulasakara, H.; Lee, V.; Brencic, A.; Liberati, N.; Urbach, J.; Miyata, S.; Lee, D.G.; Neely, A.N.; Hyodo, M.; Hayakawa, Y.; et al. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc. Natl. Acad. Sci. USA 2006, 103, 2839–2844. [Google Scholar]
- Hickman, J.W.; Harwood, C.S. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol. Microbiol 2008, 69, 376–389. [Google Scholar]
- Lee, V.T.; Matewish, J.M.; Kessler, J.L.; Hyodo, M.; Hayakawa, Y.; Lory, S. A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol. Microbiol 2007, 65, 1474–1484. [Google Scholar]
- Remminghorst, U.; Rehm, B.H. Alg44, a unique protein required for alginate biosynthesis in Pseudomonas aeruginosa. FEBS Lett 2006, 580, 3883–3888. [Google Scholar]
- Alm, R.A.; Bodero, A.J.; Free, P.D.; Mattick, J.S. Identification of a novel gene, pilZ, essential for type 4 fimbrial biogenesis in Pseudomonas aeruginosa. J. Bacteriol 1996, 178, 46–53. [Google Scholar]
- D’Argenio, D.A.; Calfee, M.W.; Rainey, P.B.; Pesci, E.C. Autolysis and autoaggregation in Pseudomonas aeruginosa colony morphology mutants. J. Bacteriol 2002, 184, 6481–6489. [Google Scholar]
- Hickman, J.W.; Tifrea, D.F.; Harwood, C.S. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl. Acad. Sci. USA 2005, 102, 14422–14427. [Google Scholar]
- Kuchma, S.L.; Brothers, K.M.; Merritt, J.H.; Liberati, N.T.; Ausubel, F.M.; O’Toole, G.A. BifA, a cyclic-Di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J. Bacteriol 2007, 189, 8165–8178. [Google Scholar]
- Meissner, A.; Wild, V.; Simm, R.; Rohde, M.; Erck, C.; Bredenbruch, F.; Morr, M.; Romling, U.; Haussler, S. Pseudomonas aeruginosa cupA-encoded fimbriae expression is regulated by a GGDEF and EAL domain-dependent modulation of the intracellular level of cyclic diguanylate. Environ. Microbiol 2007, 9, 2475–2485. [Google Scholar]
- Kirisits, M.J.; Prost, L.; Starkey, M.; Parsek, M.R. Characterization of colony morphology variants isolated from Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol 2005, 71, 4809–4821. [Google Scholar]
- Starkey, M.; Hickman, J.H.; Ma, L.; Zhang, N.; De Long, S.; Hinz, A.; Palacios, S.; Manoil, C.; Kirisits, M.J.; Starner, T.D.; et al. Pseudomonas aeruginosa rugose small-colony variants have adaptations that likely promote persistence in the cystic fibrosis lung. J. Bacteriol 2009, 191, 3492–3503. [Google Scholar]
- Malone, J.G.; Jaeger, T.; Spangler, C.; Ritz, D.; Spang, A.; Arrieumerlou, C.; Kaever, V.; Landmann, R.; Jenal, U. YfiBNR mediates cyclic di-GMP dependent small colony variant formation and persistence in Pseudomonas aeruginosa. PLoS Pathog 2010, 6, e1000804. [Google Scholar]
- Choy, W.K.; Zhou, L.; Syn, C.K.; Zhang, L.H.; Swarup, S. MorA defines a new class of regulators affecting flagellar development and biofilm formation in diverse Pseudomonas species. J. Bacteriol 2004, 186, 7221–7228. [Google Scholar]
- Kazmierczak, B.I.; Lebron, M.B.; Murray, T.S. Analysis of FimX, a phosphodiesterase that governs twitching motility in Pseudomonas aeruginosa. Mol. Microbiol 2006, 60, 1026–1043. [Google Scholar]
- Klebensberger, J.; Birkenmaier, A.; Geffers, R.; Kjelleberg, S.; Philipp, B. SiaA and SiaD are essential for inducing autoaggregation as a specific response to detergent stress in Pseudomonas aeruginosa. Environ. Microbiol 2009, 11, 3073–3086. [Google Scholar]
- Merritt, J.H.; Brothers, K.M.; Kuchma, S.L.; O’Toole, G.A. SadC reciprocally influences biofilm formation and swarming motility via modulation of exopolysaccharide production and flagellar function. J. Bacteriol 2007, 189, 8154–8164. [Google Scholar]
- Roy, A.B.; Petrova, O.E.; Sauer, K. The phosphodiesterase DipA (PA5017) is essential for Pseudomonas aeruginosa biofilm dispersion. J. Bacteriol 2012, 194, 2904–2915. [Google Scholar]
- Merritt, J.H.; Ha, D.G.; Cowles, K.N.; Lu, W.; Morales, D.K.; Rabinowitz, J.; Gitai, Z.; O’Toole, G.A. Specific control of Pseudomonas aeruginosa surface-associated behaviors by two c-di-GMP diguanylate cyclases. mBiosphere 2010, 1. [Google Scholar] [CrossRef]
- Ventre, I.; Goodman, A.L.; Vallet-Gely, I.; Vasseur, P.; Soscia, C.; Molin, S.; Bleves, S.; Lazdunski, A.; Lory, S.; Filloux, A. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc. Natl. Acad. Sci. USA 2006, 103, 171–176. [Google Scholar]
- Goodman, A.L.; Kulasekara, B.; Rietsch, A.; Boyd, D.; Smith, R.S.; Lory, S. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev. cell 2004, 7, 745–754. [Google Scholar]
- Brencic, A.; McFarland, K.A.; McManus, H.R.; Castang, S.; Mogno, I.; Dove, S.L.; Lory, S. The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the Rsm Y and RsmZ regulatory small RNAs. Mol. Microbiol 2009, 73, 434–445. [Google Scholar]
- Brencic, A.; Lory, S. Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol. Microbiol 2009, 72, 612–632. [Google Scholar]
- Irie, Y.; Starkey, M.; Edwards, A.N.; Wozniak, D.J.; Romeo, T.; Parsek, M.R. Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by Rpo S and post-transcriptionally by RsmA. Mol. Microbiol 2010, 78, 158–172. [Google Scholar]
- Goodman, A.L.; Merighi, M.; Hyodo, M.; Ventre, I.; Filloux, A.; Lory, S. Direct interaction between sensor kinase proteins mediates acute and chronic disease phenotypes in a bacterial pathogen. Genes Dev 2009, 23, 249–259. [Google Scholar]
- Kong, W.; Chen, L.; Zhao, J.; Shen, T.; Surette, M.G.; Shen, L.; Duan, K. Hybrid sensor kinase PA1611 in Pseudomonas aeruginosa regulates transitions between acute and chronic infection through direct interaction with RetS. Mol. Microbiol 2013, 88, 784–797. [Google Scholar]
- Mikkelsen, H.; McMullan, R.; Filloux, A. The Pseudomonas aeruginosa reference strain PA14 displays increased virulence due to a mutation in ladS. PLoS One 2011, 6, e29113. [Google Scholar]
- Bordi, C.; Lamy, M.C.; Ventre, I.; Termine, E.; Hachani, A.; Fillet, S.; Roche, B.; Bleves, S.; Mejean, V.; Lazdunski, A.; et al. Regulatory RNAs and the HptB/RetS signalling pathways fine-tune Pseudomonas aeruginosa pathogenesis. Mol. Microbiol 2010, 76, 1427–1443. [Google Scholar]
- Jimenez, P.N.; Koch, G.; Thompson, J.A.; Xavier, K.B.; Cool, R.H.; Quax, W.J. The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiol. Mol. Biol Rev 2012, 76, 46–65. [Google Scholar]
- Camilli, A.; Bassler, B.L. Bacterial small-molecule signaling pathways. Science 2006, 311, 1113–1116. [Google Scholar]
- Fuqua, C.; Winans, S.C.; Greenberg, E.P. Census and consensus in bacterial ecosystems: The LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu. Rev. Microbiol 1996, 50, 727–751. [Google Scholar]
- Waters, C.M.; Bassler, B.L. Quorum sensing: Cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Bio 2005, 21, 319–346. [Google Scholar]
- Schuster, M.; Greenberg, E.P. A network of networks: Quorum-sensing gene regulation in Pseudomonas aeruginosa. Int. J. Med. Microbiol 2006, 296, 73–81. [Google Scholar]
- Venturi, V. Regulation of quorum sensing in Pseudomonas. FEMS Microbiol. Rev 2006, 30, 274–291. [Google Scholar]
- De Kievit, T.R.; Gillis, R.; Marx, S.; Brown, C.; Iglewski, B.H. Quorum-sensing genes in Pseudomonas aeruginosa biofilms: Their role and expression patterns. Appl. Environ.Microbiol 2001, 67, 1865–1873. [Google Scholar]
- Sauer, K.; Camper, A.K.; Ehrlich, G.D.; Costerton, J.W.; Davies, D.G. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol 2002, 184, 1140–1154. [Google Scholar]
- Yoon, S.S.; Hennigan, R.F.; Hilliard, G.M.; Ochsner, U.A.; Parvatiyar, K.; Kamani, M.C.; Allen, H.L.; DeKievit, T.R.; Gardner, P.R.; Schwab, U.; et al. Pseudomonas aeruginosa anaerobic respiration in biofilms: Relationships to cystic fibrosis pathogenesis. Dev. Cell 2002, 3, 593–603. [Google Scholar]
- Singh, P.K.; Schaefer, A.L.; Parsek, M.R.; Moninger, T.O.; Welsh, M.J.; Greenberg, E.P. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 2000, 407, 762–764. [Google Scholar]
- Ueda, A.; Wood, T.K. Connecting quorum sensing, c-di-GMP, pel polysaccharide, and biofilm formation in Pseudomonas aeruginosa through tyrosine phosphatase TpbA (PA3885). PLoS Pathog 2009, 5, e1000483. [Google Scholar]
- Gilbert, K.B.; Kim, T.H.; Gupta, R.; Greenberg, E.P.; Schuster, M. Global position analysis of the Pseudomonas aeruginosa quorum-sensing transcription factor LasR. Mol. Microbiol 2009, 73, 1072–1085. [Google Scholar]
- Ma, L.; Wang, J.; Wang, S.; Anderson, E.M.; Lam, J.S.; Parsek, M.R.; Wozniak, D.J. Synthesis of multiple Pseudomonas aeruginosa biofilm matrix exopolysaccharides is post-transcriptionally regulated. Environ. Microbiol 2012, 14, 1995–2005. [Google Scholar]
- Tart, A.H.; Blanks, M.J.; Wozniak, D.J. The AlgT-dependent transcriptional regulator AmrZ (AlgZ) inhibits flagellum biosynthesis in mucoid, nonmotile Pseudomonas aeruginosa cystic fibrosis isolates. J. Bacteriol 2006, 188, 6483–6489. [Google Scholar]
- Jones, C.J.; Ryder, C.R.; Mann, E.E.; Wozniak, D.J. AmrZ modulates Pseudomonas aeruginosa biofilm architecture by directly repressing transcription of the psl operon. J. Bacteriol 2013, 195, 1637–1644. [Google Scholar]
- Alkawash, M.A.; Soothill, J.S.; Schiller, N.L. Alginate lyase enhances antibiotic killing of mucoid Pseudomonas aeruginosa in biofilms. Acta Pathol. Microbiol. Immunol. Scand 2006, 114, 131–138. [Google Scholar]
- Hoffmann, N.; Lee, B.; Hentzer, M.; Rasmussen, T.B.; Song, Z.; Johansen, H.K.; Givskov, M.; Hoiby, N. Azithromycin blocks quorum sensing and alginate polymer formation and increases the sensitivity to serum and stationary-growth-phase killing of Pseudomonas aeruginosa and attenuates chronic P. aeruginosa lung infection in Cftr−/− mice. Antimicrobial. Agents Chemother 2007, 51, 3677–3687. [Google Scholar]
- Florescu, D.F.; Murphy, P.J.; Kalil, A.C. Effects of prolonged use of azithromycin in patients with cystic fibrosis: A meta-analysis. Pulm. Pharmaco. Ther 2009, 22, 467–472. [Google Scholar]
- Hentzer, M.; Riedel, K.; Rasmussen, T.B.; Heydorn, A.; Andersen, J.B.; Parsek, M.R.; Rice, S.A.; Eberl, L.; Molin, S.; Hoiby, N.; et al. Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound. Microbiology 2002, 148, 87–102. [Google Scholar]
- Givskov, M.; de Nys, R.; Manefield, M.; Gram, L.; Maximilien, R.; Eberl, L.; Molin, S.; Steinberg, P.D.; Kjelleberg, S. Eukaryotic interference with homoserine lactone-mediated prokaryotic signalling. J. Bacteriol 1996, 178, 6618–6622. [Google Scholar]
- Singh, P.K.; Parsek, M.R.; Greenberg, E.P.; Welsh, M.J. A component of innate immunity prevents bacterial biofilm development. Nature 2002, 417, 552–555. [Google Scholar]
- Musk, D.J.; Banko, D.A.; Hergenrother, P.J. Iron salts perturb biofilm formation and disrupt existing biofilms of Pseudomonas aeruginosa. Chem. Biol 2005, 12, 789–796. [Google Scholar]
- Zeng, X.; Liu, X.; Bian, J.; Pei, G.; Dai, H.; Polyak, S.W.; Song, F.; Ma, L.; Wang, Y.; Zhang, L. Synergistic effect of 14-alpha-lipoyl andrographolide and various antibiotics on the formation of biofilms and production of exopolysaccharide and pyocyanin by Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2011, 55, 3015–3017. [Google Scholar]
- Kolodkin-Gal, I.; Romero, D.; Cao, S.; Clardy, J.; Kolter, R.; Losick, R. d-amino acids trigger biofilm disassembly. Science 2010, 328, 627–629. [Google Scholar]
- Kolodkin-Gal, I.; Cao, S.; Chai, L.; Bottcher, T.; Kolter, R.; Clardy, J.; Losick, R. A self-produced trigger for biofilm disassembly that targets exopolysaccharide. Cell 2012, 149, 684–692. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Wei, Q.; Ma, L.Z. Biofilm Matrix and Its Regulation in Pseudomonas aeruginosa. Int. J. Mol. Sci. 2013, 14, 20983-21005. https://doi.org/10.3390/ijms141020983
Wei Q, Ma LZ. Biofilm Matrix and Its Regulation in Pseudomonas aeruginosa. International Journal of Molecular Sciences. 2013; 14(10):20983-21005. https://doi.org/10.3390/ijms141020983
Chicago/Turabian StyleWei, Qing, and Luyan Z. Ma. 2013. "Biofilm Matrix and Its Regulation in Pseudomonas aeruginosa" International Journal of Molecular Sciences 14, no. 10: 20983-21005. https://doi.org/10.3390/ijms141020983




