A Comparative Proteomic Analysis of Pinellia ternata Leaves Exposed to Heat Stress
Abstract
:1. Introduction
2. Results
2.1. Physiological Responses Induced by Heat Stress
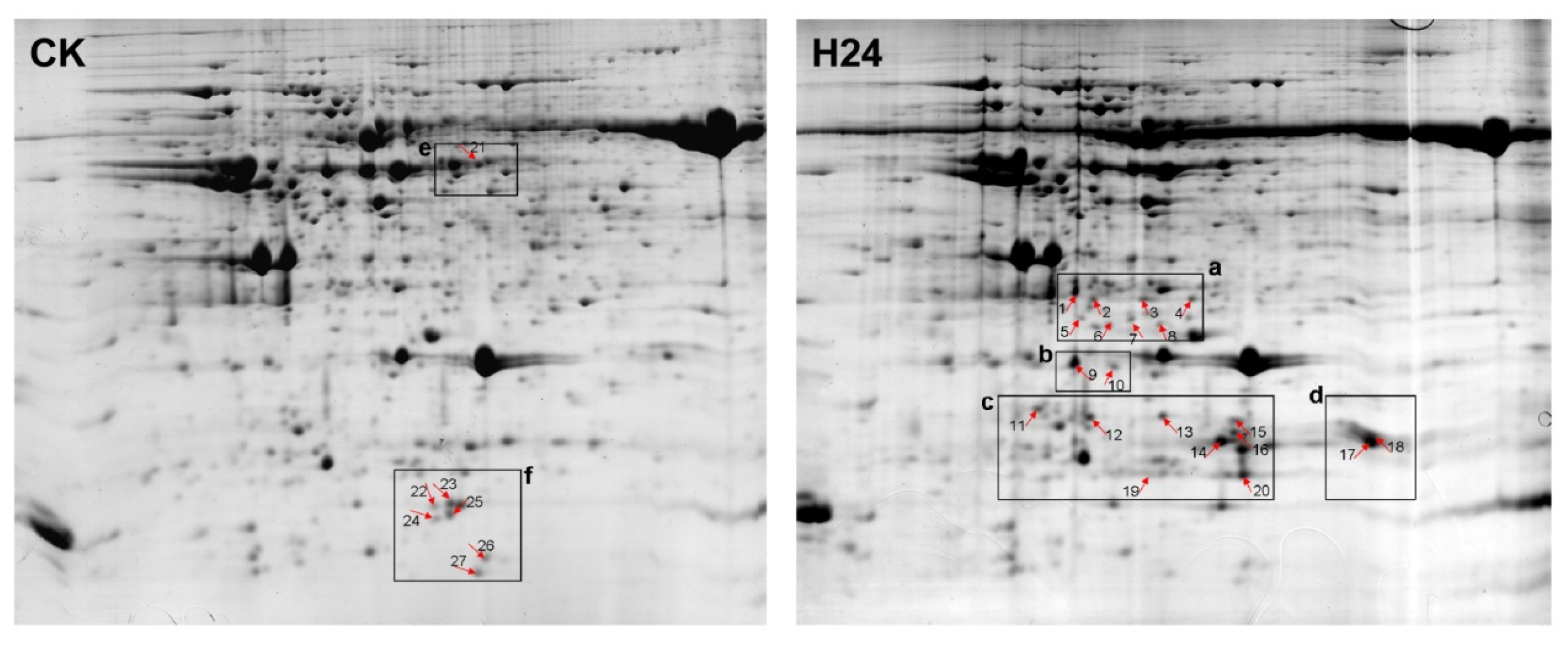
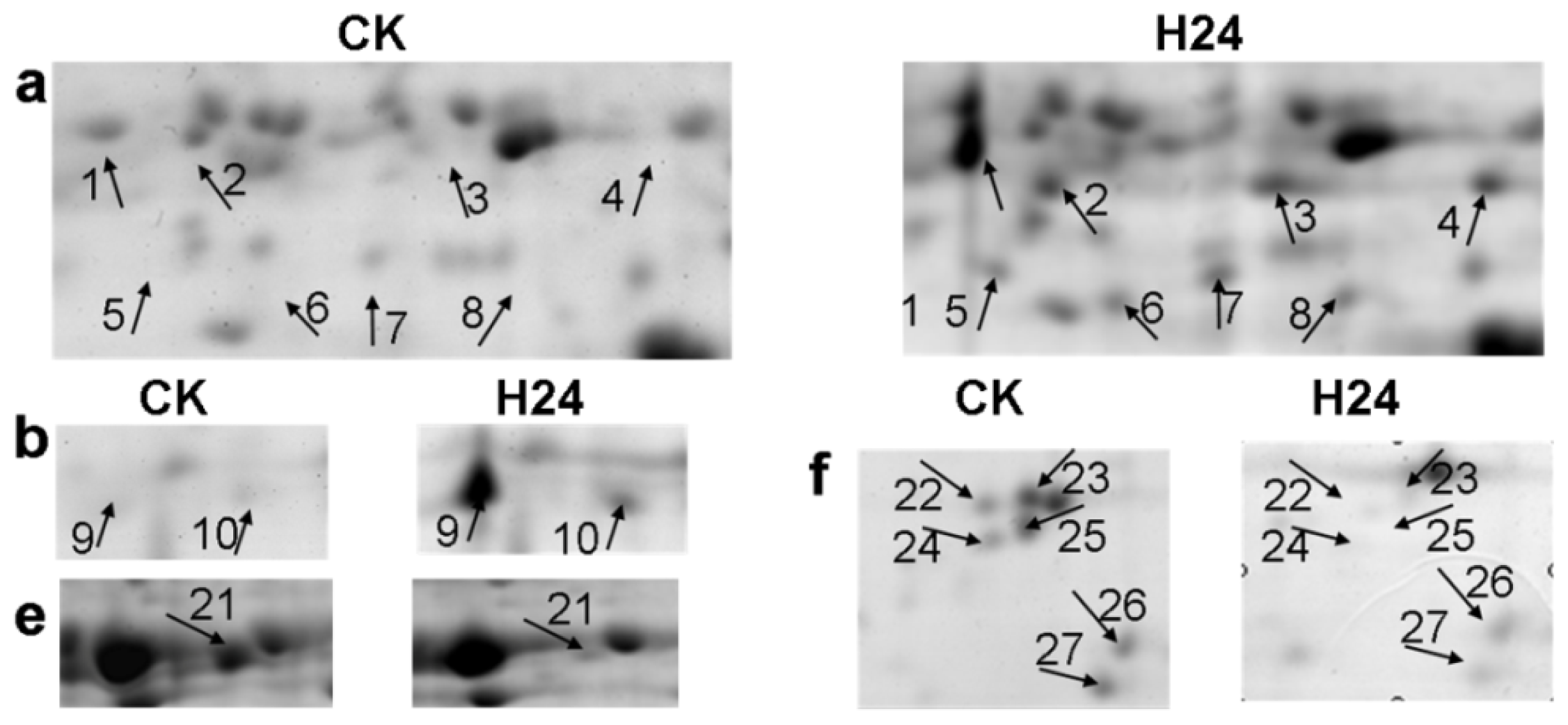
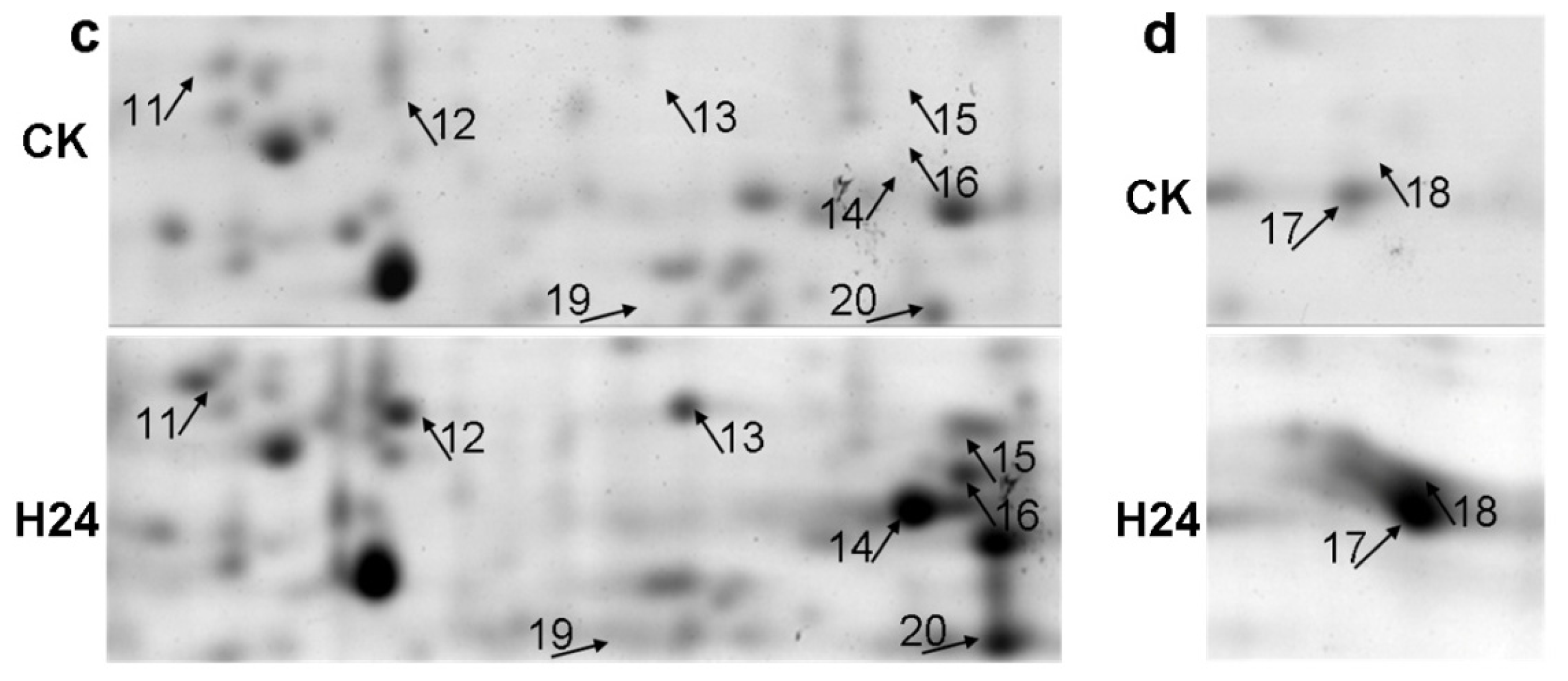
2.2. 2-DE Analysis of P. ternata Leaf Proteins Expressed in Response to Heat Stress
2.3. Identification of Heat-Responsive Proteins
2.4. Gene Expression Analysis by qPCR
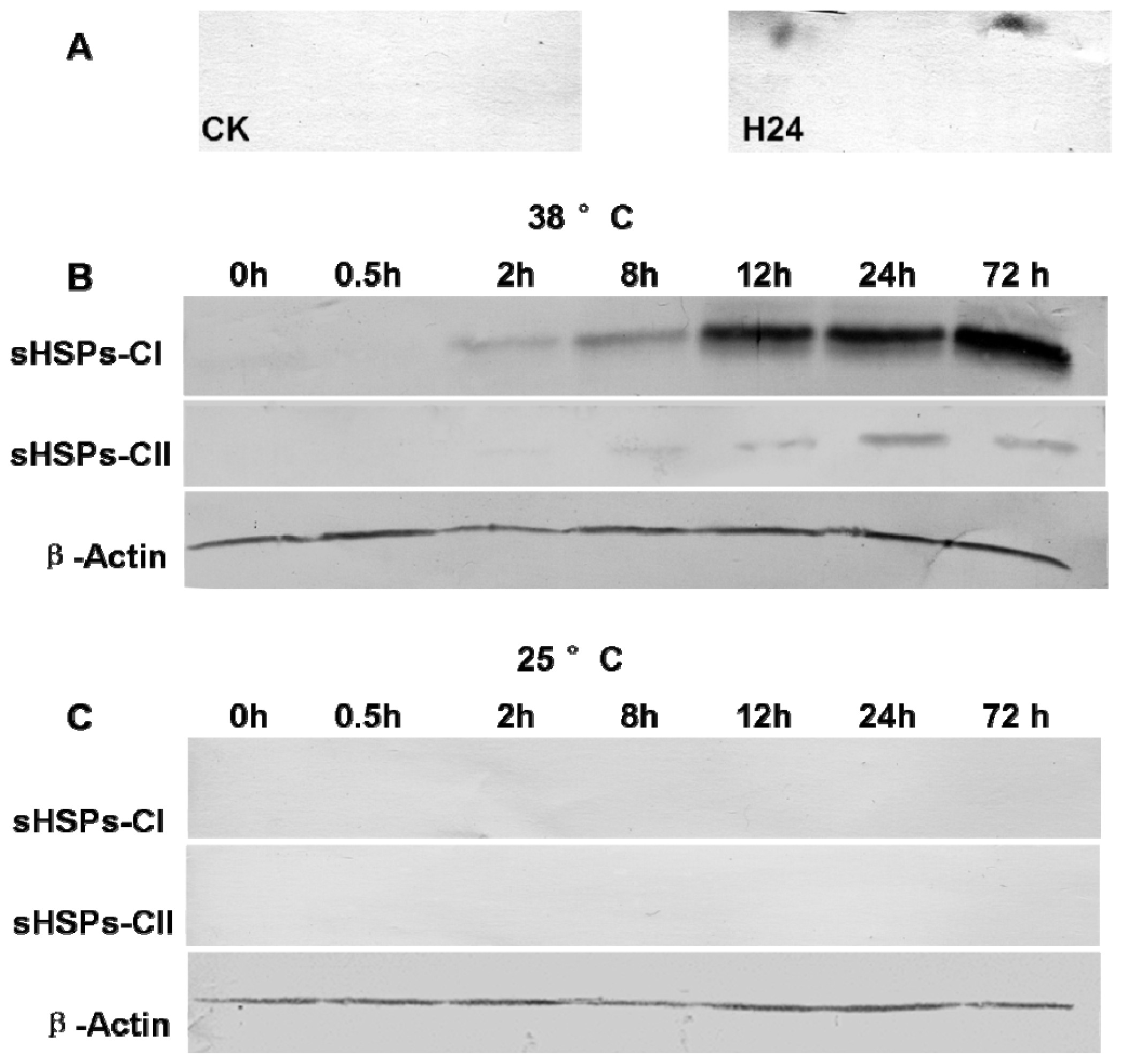
2.5. Western Blot Analysis of sHSPs-CI and sHSPs-CII
3. Discussion
3.1. Photosynthesis-Related Proteins
3.2. Small Heat Shock Proteins
3.3. Chlorophyll Biosynthetic Process Proteins
3.4. Protein Degradation Proteins
3.5. RNA Processing Proteins
3.6. Defense Proteins
4. Experimental Section
4.1. Plant Materials and Treatment
4.2. Physiological Parameter Measurement
4.3. Protein Extraction
4.4. Two-Dimensional Gel Electrophoresis and Image Analysis
4.5. Tandem Mass Spectrometry and Database Searches
4.6. RNA Isolation and Real-Time Quantitative PCR (qPCR)
4.7. Western Blot Analysis
5. Conclusions
Supplementary Information
ijms-14-20614-s001.pdf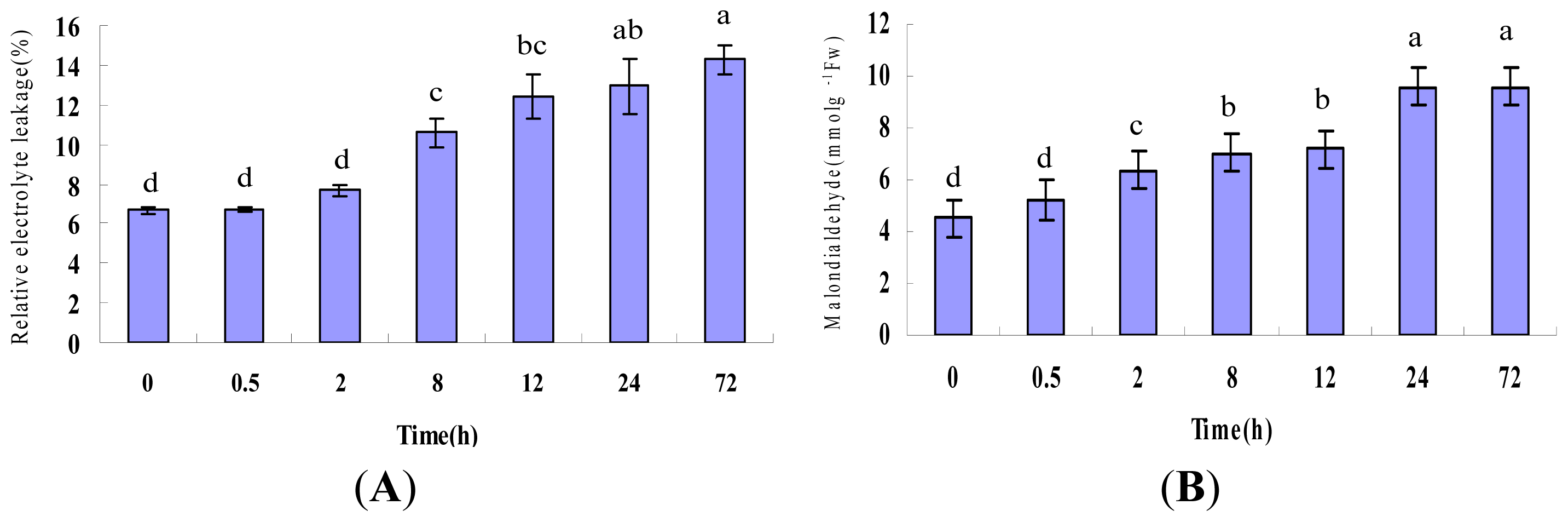
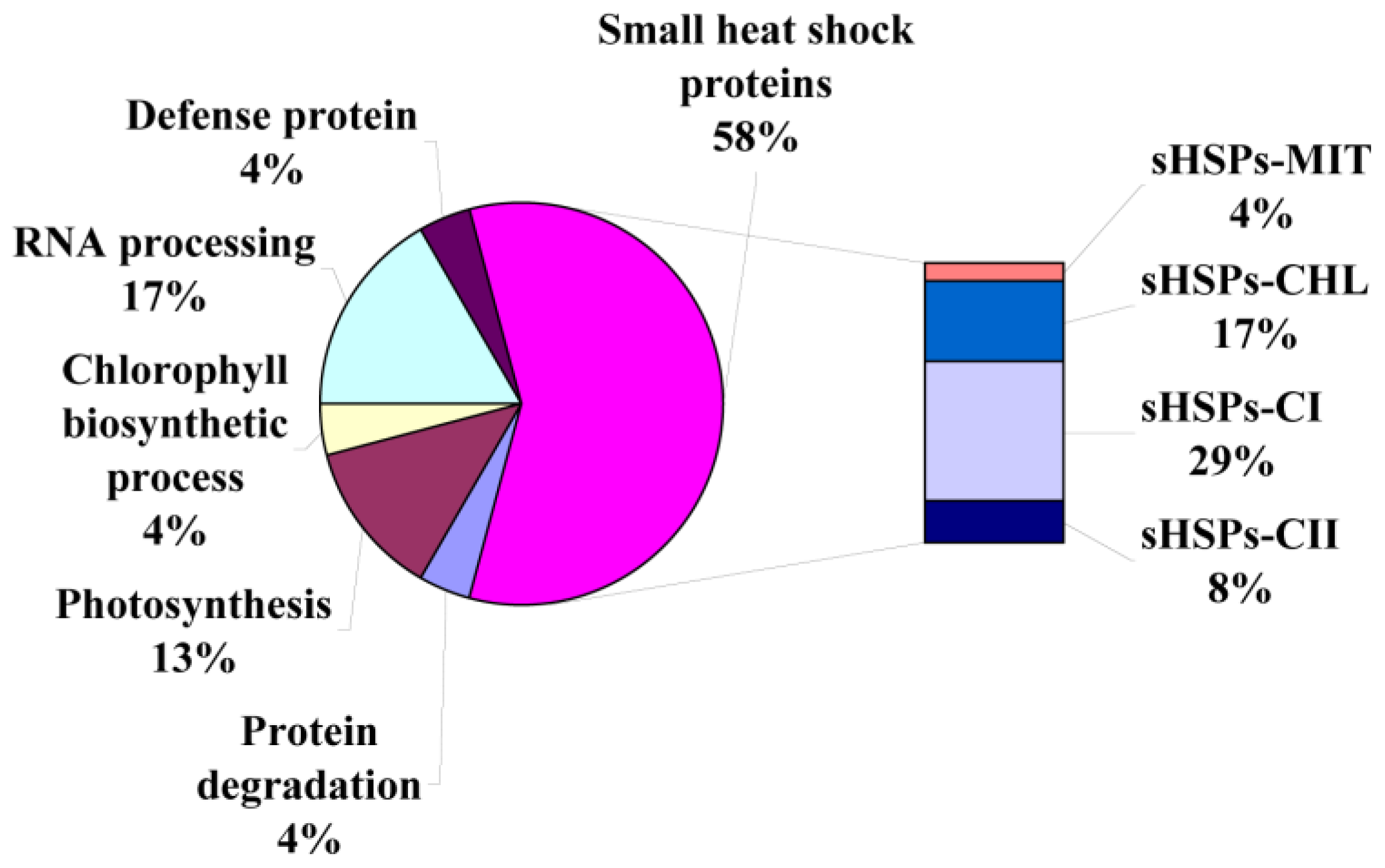

| Spot No. | Protein Name | Accession No. | Organism | Theor. Mr (Da)/pI | Exper. Mr (Da)/pI | Coverage (%) | Score | Subcellular localization | Matched peptides |
|---|---|---|---|---|---|---|---|---|---|
| Photosynthesis | |||||||||
| 1 | chlorophyll a/b-binding protein | EMS50798 | Triticum urartu | 31,970/5.75 | 31,436/5.02 | 6 | 128 | chloroplast | 3 |
| 2 | chlorophyll a/b-binding protein | EMS50798 | Triticum urartu | 31,970/5.75 | 30,489/5.09 | 6 | 128 | chloroplast | 3 |
| 18 | Rieske Fe/S protein of cytochrome b6/f complex | CAA45705 | Nicotiana tabacum | 24,511/7.59 | 17,911//6.11 | 5 | 68 | chloroplast | 1 |
| Small heat shock proteins | |||||||||
| 3 | small heat shock protein | AEX97053 | Copaifera officinalis | 27,275/6.97 | 31,012/5.29 | 4 | 89 | chloroplast | 1 |
| 4 | small heat shock protein | AEX97053 | Copaifera officinalis | 27,275/6.97 | 31,151/5.38 | 4 | 89 | chloroplast | 1 |
| 6 | small heat shock protein | AEX97053 | Copaifera officinalis | 27,275/6.97 | 27,867/5.14 | 4 | 54 | chloroplast | 2 |
| 8 | small heat shock protein | AEX97053 | Copaifera officinalis | 27,275/6.97 | 28,714/5.34 | 4 | 54 | chloroplast | 1 |
| 10 | mitochondrial shsp | NP_001233872.1 | Solanum lycopersicum | 23,833/6.45 | 24,203/5.17 | 9 | 63 | mitochondrial | 2 |
| 11 | HSP21 | ABW81098 | Cleome spinosa | 17,432/5.97 | 18,341/4.88 | 11 | 108 | cytoplasm | 3 |
| 12 | 17.5 kDa class I hsp | AAR25848 | Carica papaya | 17,471/5.31 | 18,274/5.08 | 11 | 72 | cytoplasm | 2 |
| 13 | heat shock protein 17.9 | CAA63903 | Cenchrus americanus | 17,935/5.82 | 18,282/5.34 | 9 | 107 | cytoplasm | 3 |
| 14 | 18.2 kDa class I hsp | P27880 | Medicago sativa | 18,154/5.81 | 17,845/5.55 | 31 | 403 | cytoplasm | 4 |
| 15 | heat shock protein 17.9 | CAA63903 | Cenchrus americanus | 17,935/5.82 | 18,065/5.60 | 9 | 107 | cytoplasm | 3 |
| 16 | 18.2 kDa class I hsp | XP_002281220 | Vitis vinifera | 17,041/5.80 | 17,901/5.60 | 11 | 100 | cytoplasm | 2 |
| 17 | 17.7 kDa heat shock protein | AAB63311 | Helianthus annuus | 17,662/6.19 | 17,839/6.11 | 17 | 175 | cytoplasm | 3 |
| 19 | small heat shock protein | AFK28040 | Pinellia ternata | 17,231/5.52 | 17,346/5.30 | 10 | 62 | cytoplasm | 1 |
| 20 | small heat shock protein | AFK28040 | Pinellia ternata | 17,231/5.52 | 17,346/5.64 | 10 | 62 | cytoplasm | 1 |
| Chlorophyll biosynthetic process | |||||||||
| 21 | Glutamate-1-semialdehyde 2,1-aminomutase | GSA_HORVU | Hordeum vulgare | 49,690/6.39 | 48,096/5.84 | 5 | 62 | chloroplast | 2 |
| Protein degradation | |||||||||
| 9 | putative spop | BAC66712 | Oryza sativa Japonica Group | 41,986/5.32 | 24,722/5.04 | 4 | 52 | cytoplasm | 1 |
| RNA processing | |||||||||
| 22 | glycine-rich RNA-binding protein | XP_003631658 | Vitis vinifera | 16,376/6.32 | 14,987/5.62 | 19 | 62 | nucleus | 2 |
| 23 | glycine-rich RNA-binding protein | ACJ11730 | Gossypium hirsutum | 17,075/7.82 | 15,011/5.67 | 16 | 204 | nucleus | 4 |
| 24 | glycine-rich RNA-binding protein GRP1A-like | XP_003631658 | Vitis vinifera | 16,376/6.32 | 14,946/5.62 | 19 | 198 | nucleus | 4 |
| 25 | glycine-rich RNA-binding protein | AAT85299 | Oryza sativa | 15,949/6.29 | 14,963/5.67 | 8 | 79 | nucleus | 2 |
| Defense protein | |||||||||
| 26 | lectin | AAP20876 | Pinellia ternata | 29,720/6.58 | 11,530/5.79 | 11 | 180 | extracellular | 4 |
| Gene Name | Sequence | |
|---|---|---|
| sHSP-CII | -F | 5′-CGATCACACAGCACTCAACAAC-3′ |
| -R | 5′-GCTCTCCAGTCCCAACATCC-3′ | |
| GRP | -F | 5′-CTTCGGCTTCGTGACCTTT-3′ |
| -R | 5′-GGTTGACGGTGATGCTCCT-3′ | |
| sHSP-MIT | -F | 5′-GGTGGAGCAGAACACTTTGG-3′ |
| -R | 5′-GCACCCCGTTCTTCATCTC-3′ | |
| GAPDH | -F | 5′-TGCTGGGAATGATGTTGAATG-3′ |
| -R | 5′-TTGGCATTGTTGAGGGTTTG-3′ | |
| sHSP-CI | -F | 5′-GACTCGGAGACCTCTGCTTT-3′ |
| -R | 5′-TCACTTCCTCCTTGTTGACG-3′ | |
Acknowledgments
Conflicts of Interest
References
- Wahid, A.; Gelani, S.; Ashraf, M.; Foolad, M.R. Heat tolerance in plants: An overview. Environ. Exp. Bot 2007, 61, 199–223. [Google Scholar]
- Wang, W.X.; Vinocur, B.; Shoseyov, O.; Altman, A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci 2004, 9, 244–252. [Google Scholar]
- Vierling, E. The roles of heat shock proteins in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol 1991, 42, 579–620. [Google Scholar]
- Boston, R.S.; Viitanen, P.V.; Vierling, E. Molecular chaperones and protein folding in plants. Plant Mol. Biol 1996, 32, 191–222. [Google Scholar]
- Wang, W.X.; Vinocur, B.; Altman, A. Plant responses to drought, salinity and extreme temperatures: Towards genetic engineering for stress tolerance. Planta 2003, 218, 1–14. [Google Scholar]
- Lin, S.K.; Chang, M.C.; Tsai, Y.G.; Lur, H.S. Proteomic analysis of the expression of proteins related to rice quality during caryopsis development and the effect of high temperature on expression. Proteomics 2005, 5, 2140–2156. [Google Scholar]
- Lee, D.G.; Ahsan, N.; Lee, S.H.; Kang, K.Y.; Bahk, J.D.; Lee, I.J.; Lee, B.H. A proteomic approach in analyzing heat-responsive proteins in rice leaves. Proteomics 2007, 7, 3369–3383. [Google Scholar]
- Skylasa, D.J.; Cordwell, S.; Hains, P.G.; Larsena, M.R.; Basseala, D.J.; Walsha, B.J.; Blumenthal, C.; Rathmell, W.; Copeland, L.; Wrigley, C.W. Heat shock of wheat during grain filling: Proteins associated with heat tolerance. J. Cereal Sci 2002, 35, 175–188. [Google Scholar]
- Majoul, T.; Bancel, E.; Triboi, E.; Ben Hamida, J.; Branlard, G. Proteomic analysis of the effect of heat stress on hexaploid wheat grain: Characterization of heat-responsive proteins from total endosperm. Proteomics 2003, 3, 175–183. [Google Scholar]
- Laino, P.; Shelton, D.; Finnie, C.; de Leonardis, A.M.; Mastrangelo, A.M.; Svensson, B.; Lafiandra, D.; Masci, S. Comparative proteome analysis of metabolic proteins from seeds of durum wheat (cv. Svevo) subjected to heat stress. Proteomics 2010, 10, 2359–2368. [Google Scholar] [Green Version]
- Sule, A.; Vanrobaeys, F.; Hajos, G.; van Beeumen, J.; Devreese, B. Proteomic analysis of small heat shock protein isoforms in barley shoots. Phytochemistry 2004, 65, 1853–1863. [Google Scholar]
- Hu, X.L.; Li, Y.H.; Li, C.H.; Yang, H.R.; Wang, W.; Lu, M.H. Characterization of small heat shock proteins associated with maize tolerance to combined drought and heat stress. J. Plant Growth Regul 2010, 29, 455–464. [Google Scholar]
- Liu, T.; Zhang, L.; Yuan, Z.; Hu, X.; Lu, M.; Wang, W.; Wang, Y. Identification of proteins regulated by ABA in response to combined drought and heat stress in maize roots. Acta Physiol. Plant 2013, 35, 501–513. [Google Scholar]
- Kosova, K.; Vitamvas, P.; Prasil, I.T.; Renaut, J. Plant proteome changes under abiotic stress—Contribution of proteomics studies to understanding plant stress response. J. Proteomics 2011, 74, 1301–1322. [Google Scholar]
- Hasanuzzaman, M.; Nahar, K.; Alam, M.M.; Roychowdhury, R.; Fujita, M. Physiological, biochemical, and molecular mechanisms of heat stress tolerance in plants. Int. J. Mol. Sci 2013, 14, 9643–9684. [Google Scholar]
- Chen, J.H.; Cui, G.Y.; Liu, J.Y.; Tan, R.X. Pinelloside, an antimicrobial cerebroside from Pinellia ternata. Phytochemistry 2003, 64, 903–906. [Google Scholar]
- Kim, Y.; Shin, Y.; Ha, Y.; Lee, S.; Oh, J.; Kim, Y.S. Anti-obesity effect of Pinellia ternate extract in Zucker rats. Biol. Pharm. Bull 2006, 29, 1278–1281. [Google Scholar]
- Guo, Q.; Zhang, G.; Wang, K. Studies on ecological prerequisites for growth of Pinellia ternate. Acta Bot. Yunnanica 1998, Suppl, 67–70. [Google Scholar]
- Zhu, Y.H.; Zhu, G.S.; Zhu, Z.B.; Guo, Q.S.; Liu, Z.Y. Cloning of a cytoplasmic small heat shock protein gene and expression analysis in Pinellia ternata. Plant Cell Rep 2013.
- Higashi, Y.; Hirai, M.Y.; Fujiwara, T.; Naito, S.; Noji, M.; Saito, K. Proteomic and transcriptomic analysis of Arabidopsis seeds: Molecular evidence for successive processing of seed proteins and its implication in the stress response to sulfur nutrition. Plant J 2006, 48, 557–571. [Google Scholar]
- Xue, J.P.; Zhang, A.M.; Yang, J.; Chang, L.; Huang, Y.Q. Change of endogenous hormone around sprout tumble of Pinellia ternata under high temperature stress. Zhongguo Zhong Yao Za Zhi 2007, 32, 2489–2491. [Google Scholar]
- Lu, H.; Xue, T.; Zhang, A.; Sheng, W.; Zhu, Y.; Chang, L.; Song, Y.; Xue, I. Construction of an SSH library of Pinellia ternate under heat stress, and expression analysis of four transcripts. Plant Mol. Biol. Rep 2013, 31, 185–194. [Google Scholar]
- Carpentier, S.C.; Panis, B.; Vertommen, A.; Swennen, R.; Sergeant, K.; Renaut, J.; Laukens, K.; Witters, E.; Samyn, B.; Devreese, B. Proteome analysis of non-model plants: A challenging but powerful approach. Mass Spectrom. Rev 2008, 27, 354–377. [Google Scholar]
- Xiong, E.; Wu, X.; Shi, J.; Wang, X.; Wang, W. Proteomic identification of differentially expressed proteins between male and female plants in Pistacia chinensis. PLoS One 2013, 8, e64276. [Google Scholar]
- Samyn, B.; Sergeant, K.; Carpentier, S.; Debyser, G.; Panis, B.; Swennen, R.; van Beeumen, J. Functional proteome analysis of the banana plant (Musa spp.) using de novo sequence analysis of derivatized peptides. J. Proteome Res 2007, 6, 70–80. [Google Scholar]
- Plomion, C.; Lalanne, C.; Claverol, S.; Meddour, H.; Kohler, A.; Bogeat-Triboulot, M.B.; Barre, A.; le Provost, G.; Dumazet, H.; Jacob, D.; et al. Mapping the proteome of poplar and application to the discovery of drought-stress responsive proteins. Proteomics 2006, 6, 6509–6527. [Google Scholar]
- Xu, C.P.; Huang, B.R. Comparative analysis of proteomic responses to single and simultaneous drought and heat stress for two kentucky bluegrass cultivars. Crop. Sci 2012, 52, 1246–1260. [Google Scholar]
- Waters, E.R. The molecular evolution of the small heat-shock proteins in plants. Genetics 1995, 141, 785–795. [Google Scholar]
- Horton, P.; Park, K.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K; WoLF, PSORT. Protein localization predictor. Nucleic Acids Res 2007, 35, 585–587. [Google Scholar]
- Jacob, A.; Lancaster, J.; Buhler, J.; Harris, B.; Chamberlain, R.D.; Mercury, BLASTP. Accelerating protein sequence alignment. ACM Trans. Reconfig. Technol. Syst 2008, 1. [Google Scholar] [CrossRef]
- Siddique, M.; Gernhard, S.; von Koskull-Doring, P.; Vierling, E.; Scharf, K.D. The plant sHSP superfamily: Five new members in Arabidopsis thaliana with unexpected properties. Cell Stress Chaperon 2008, 13, 183–197. [Google Scholar]
- Sun, W.N.; Bernard, C.; van de Cotte, B.; van Montagu, M.; Verbruggen, N. At-HSP17.6A, encoding a small heat-shock protein in Arabidopsis, can enhance osmotolerance upon overexpression. Plant J 2001, 27, 407–415. [Google Scholar]
- Perez, D.E.; Hoyer, J.S.; Johnson, A.I.; Moody, Z.R.; Lopez, J.; Kaplinsky, N.J. Bobber1 is a noncanonical Arabidopsis small heat shock protein required for both development and thermotolerance. Plant Physiol 2009, 151, 241–252. [Google Scholar]
- Sun, L.; Liu, Y.; Kong, X.; Zhang, D.; Pan, J.; Zhou, Y.; Wang, L.; Li, D.; Yang, X. ZmHSP16.9, a cytosolic class I small heat shock protein in maize (Zea mays), confers heat tolerance in transgenic tobacco. Plant Cell Rep 2012, 31, 1473–1484. [Google Scholar]
- Charng, Y.Y.; Liu, H.C.; Liu, N.Y.; Hsu, F.C.; Ko, S.S. Arabidopsis Hsa32, a novel heat shock protein, is essential for acquired thermotolerance during long recovery after acclimation. Plant Physiol 2006, 140, 1297–1305. [Google Scholar]
- Downs, C.A.; Ryan, S.L.; Heckathorn, S.A. The chloroplast small heat-shock protein: Evidence for a general role in protecting photosystem II against oxidative stress and photoinhibition. J. Plant Physiol 1999, 155, 488–496. [Google Scholar]
- Sanmiya, K.; Suzuki, K.; Egawa, Y.; Shono, M. Mitochondrial small heat-shock protein enhances thermotolerance in tobacco plants. Febs Lett 2004, 557, 265–268. [Google Scholar]
- Downs, C.A.; Heckathorn, S.A. The mitochondrial small heat-shock protein protects NADH:ubiquinone oxidoreductase of the electron transport chain during heat stress in plants. Febs Lett 1998, 430, 246–250. [Google Scholar]
- Balmer, Y.; Koller, A.; del Val, G.; Manieri, W.; Schurmann, P.; Buchanan, B.B. Proteomics gives insight into the regulatory function of chloroplast thioredoxins. Proc. Natl. Acad. Sci. USA 2003, 100, 370–375. [Google Scholar]
- Finn, R.D.; Mistry, J.; Schuster-Bockler, B.; Griffiths-Jones, S.; Hollich, V.; Lassmann, T.; Moxon, S.; Marshall, M.; Khanna, A.; Durbin, R.; et al. Pfam: Clans, web tools and services. Nucleic Acids Res 2006, 34, 247–251. [Google Scholar]
- Weber, H.; Bernhardt, A.; Dieterle, M.; Han, P.; Hano, P.; Mutlu, A.; Estelle, M.; Genschik, P.; Hellmann, H. Arabidopsis AtCUL3a and AtCUL3b form complexes with members of the BTB/POZ-MATH protein family. Plant Physiol 2005, 137, 83–93. [Google Scholar]
- Weber, H.; Hellmann, H. Arabidopsis thaliana BTB/POZ-MATH proteins interact with members of the ERF/AP2 transcription factor family. FEBS J 2009, 276, 6624–6635. [Google Scholar]
- Lorkovic, Z.J. Role of plant RNA-binding proteins in development, stress response and genome organization. Trends Plant Sci 2009, 14, 229–236. [Google Scholar]
- Cao, S.; Jiang, L.; Song, S.; Jing, R.; Xu, G. AtGRP7 is involved in the regulation of abscisic acid and stress responses in Arabidopsis. Cell Mol. Biol. Lett 2006, 11, 526–535. [Google Scholar]
- Kim, J.S.; Jung, H.J.; Lee, H.J.; Kim, K.A.; Goh, C.H.; Woo, Y.M.; Oh, S.H.; Han, Y.S.; Kang, H. Glycine-rich RNA-binding protein7 affects abiotic stress responses by regulating stomata opening and closing in Arabidopsis thaliana. Plant J 2008, 55, 455–466. [Google Scholar]
- Wang, C.; Zhang, D.W.; Wang, Y.C.; Zheng, L.; Yang, C.P. A glycine-rich RNA-binding protein can mediate physiological responses in transgenic plants under salt stress. Mol. Biol. Rep 2012, 39, 1047–1053. [Google Scholar]
- Yao, J.; Zhao, X.; Liao, Z.; Lin, J.; Chen, Z. Cloning and molecular characterization of a novel lectin gene from Pinellia ternata. Cell Res 2003, 13, 301–308. [Google Scholar]
- Lin, J.; Yao, J.H.; Zhou, X.W.; Sun, X.F.; Tang, K.X. Expression and purification of a novel mannose-binding lectin from Pinellia ternata. Mol. Biotechnol 2003, 25, 215–221. [Google Scholar]
- Peumans, W.J.; van Damme, E.J. Lectins as plant defense proteins. Plant Physiol 1995, 109, 347–352. [Google Scholar]
- Chrispeels, M.J.; Raikhel, N.V. Lectins, lectin genes, and their role in plant defense. Plant Cell 1991, 3, 1–9. [Google Scholar]
- Yang, F.; Xiao, X.; Zhang, S.; Korpelainen, H.; Li, C. Salt stress responses in Populus cathayana Rehder. Plant Sci 2009, 176, 669–677. [Google Scholar]
- Wang, W.; Vignani, R.; Scali, M.; Cresti, M. A universal and rapid protocol for protein extraction from recalcitrant plant tissues for proteomic analysis. Electrophoresis 2006, 27, 2782–2786. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem 1976, 72, 248–254. [Google Scholar]
- Korotaeva, N.E.; Oskorbina, M.V.; Kopytova, L.D.; Suvorova, G.G.; Borovskii, G.B.; Voinikov, V.K. Variations in the content of stress proteins in the needles of common pine (Pinus sylvestris L.) within an annual cycle. J. For. Res 2012, 17, 89–97. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Zhu, Y.; Zhu, G.; Guo, Q.; Zhu, Z.; Wang, C.; Liu, Z. A Comparative Proteomic Analysis of Pinellia ternata Leaves Exposed to Heat Stress. Int. J. Mol. Sci. 2013, 14, 20614-20634. https://doi.org/10.3390/ijms141020614
Zhu Y, Zhu G, Guo Q, Zhu Z, Wang C, Liu Z. A Comparative Proteomic Analysis of Pinellia ternata Leaves Exposed to Heat Stress. International Journal of Molecular Sciences. 2013; 14(10):20614-20634. https://doi.org/10.3390/ijms141020614
Chicago/Turabian StyleZhu, Yunhao, Guosheng Zhu, Qiaosheng Guo, Zaibiao Zhu, Changlin Wang, and Zuoyi Liu. 2013. "A Comparative Proteomic Analysis of Pinellia ternata Leaves Exposed to Heat Stress" International Journal of Molecular Sciences 14, no. 10: 20614-20634. https://doi.org/10.3390/ijms141020614




