Development of New Microsatellite Markers for Salvia officinalis L. and Its Potential Use in Conservation-Genetic Studies of Narrow Endemic Salvia brachyodon Vandas
Abstract
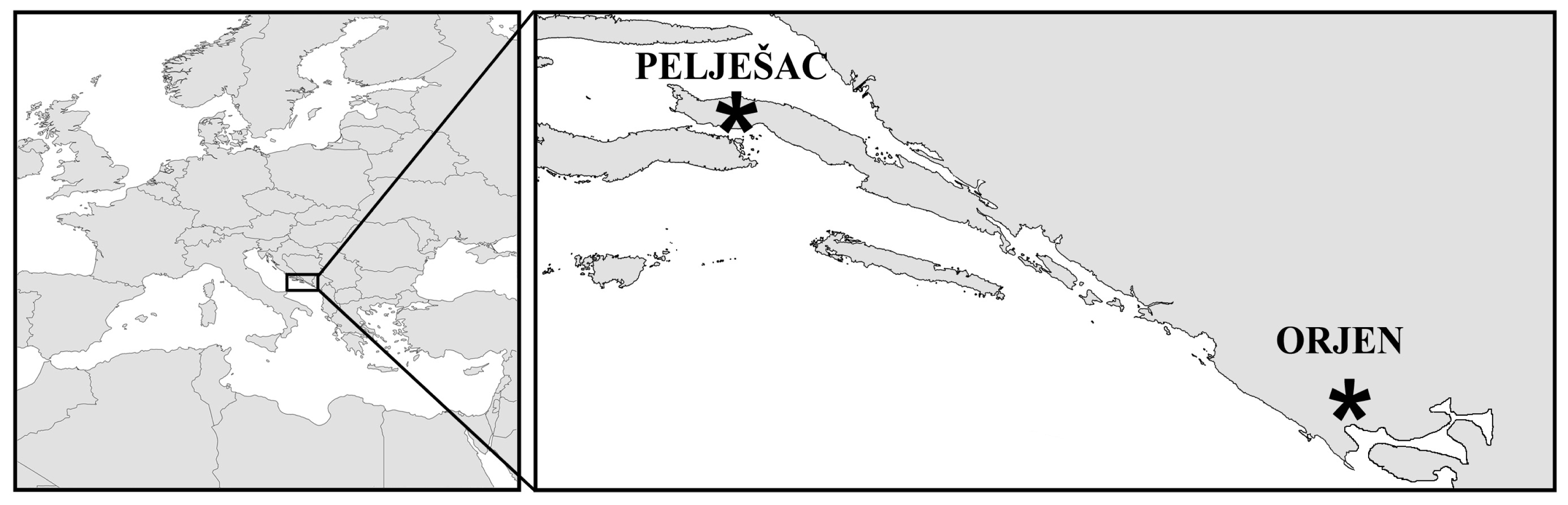
:1. Introduction
2. Results and Discussion
2.1. Development of New Microsatellite Markers for Dalmatian Sage
2.2. Cross-Amplification in Narrow Endemic Salvia brachyodon
2.3. Population Genetics Parameters and Structures of Natural Populations of Two Closely Related Species
3. Experimental Section
4. Conclusions
Acknowledgments
References
- Walker, J.B.; Sytsma, K.J.; Treutlein, J.; Wink, M. Salvia (Lamiacae) is not monophyletic: Implications for the systematics, radiation, and ecological specializations of Salvia and tribe Mentheae. Am. J. Bot 2004, 91, 1115–1125. [Google Scholar]
- Hedge, I.C. Salvia. In Flora Europaea; Tutin, T.G., Heywood, V.H., Burges, N.A., Moore, D.M., Valentine, D.H., Walters, S.M., Webb, D.A., Eds.; Cambridge University Press: Cambridge, UK, 1972; Volume 3, pp. 188–192. [Google Scholar]
- Putievsky, E.; Ravid, U.; Diwan-Rinzler, N.; Zohary, D. Genetic affinities and essential oil composition of Salvia officinalis L., S. fruticosa Mill., S. tomentosa Mill. and their hybrids. Flavour Frag. J 1990, 5, 121–123. [Google Scholar]
- Baricevic, D.; Bartol, T. The Biological/Pharmacological Activity of the Salvia Genus. In Sage the Genus Salvia; Kintzios, S.E., Ed.; Harwood Academic Publishers: Amsterdam, The Netherlands, 2000; pp. 143–184. [Google Scholar]
- Amr, S.; Đorđević, S. The investigation of the quality of Sage (Salvia officinalis) originating from Jordan. Facta Univ. Ser. Work. Living Environ. Prot 2000, 1, 103–108. [Google Scholar]
- Armitage, A.M. Herbaceous Perennial Plants: A Treatise on Their Identification, Culture, and Garden Attributes, 2nd ed; Stipes Publishing: Champaign, IL, USA, 1997; pp. 902–913. [Google Scholar]
- Reales, A.; Rivera, D.; Palazón, J.A.; Obón, C. Numerical taxonomy study of Salvia sect. Salvia (Labiatae). Bot. J. Linn. Soc 2004, 145, 353–371. [Google Scholar]
- Adamović, L. The Flora of Dalmatia (in German); Verlag von Dr. Werner Klinkhardt: Leipzig, Germany, 1911; p. 113. [Google Scholar]
- Adamović, L. The Flora of the Adriatic Countries Including Eastern Italy, Istria, Kvarner Islands of the Croatian Coast, Dalmatia, Southern Hercegovina, Southern Montenegro and Albania (in German); Verlag von Gustav Fischer: Jena, Germany, 1929. [Google Scholar]
- Girometta, M. Mount mosor (in Croatian). Hrvat. Planin 1930, 3, 76–86. [Google Scholar]
- Barbalić, Lj. Contribution to the knowledge of Salvia brachyodon Vand (in Croatian). Biol. Glas. 1956, 9, 5–10. [Google Scholar]
- Abadžić, S.; Šilić, Č. Chorology, ecology and phytosociological affiliation of Salvia brachyodon Vandas in flora of Yugoslavia (in Montenegrin). Glas. Repub. Zavoda Zašt. Prir. Prir. Muz. Titogr. 1982, 15, 125–131. [Google Scholar]
- Šilić, Č. Endemic Plants (in Bosnian); Svjetlost: Sarajevo & Beograd, Jugoslavija, 1984; p. 107. [Google Scholar]
- Savikin-Fodulovica, K.P.; Tasic, S.R.; Menkovic, N.R. The essential oil of Salvia brachyodon Vandas Lamiaceae. J. Essent. Oil Res 2002, 14, 342–343. [Google Scholar]
- Tzakou, O.; Couladis, M.; Slavkovska, V.; Mimica-Dukic, N.; Jancic, R. The essential oil composition of Salvia brachyodon Vandas. Flavour Frag. J 2003, 18, 2–4. [Google Scholar]
- Flora Croatica Database. Available online: http://hirc.botanic.hr/fcd accessed on 26 June 2012.
- Petrović, D.; Stešević, D.; Vuksanović, S. Materials for the Red Book of Montenegro. Nat. Montenegrina 2008, 7, 605–631. [Google Scholar]
- Slatkin, M. Gene flow and the geographic structure of natural populations. Science 1987, 236, 787–792. [Google Scholar]
- Schaal, B.A.; Hayworth, D.A.; Olsen, K.M.; Rauscher, J.T.; Smith, W.A. Phylogeographic studies in plants: Problems and prospects. Mol. Ecol 1998, 7, 465–474. [Google Scholar]
- Brookfield, J.F.Y. A simple new method for estimating null allele frequency from heterozygote deficiency. Mol. Ecol 1996, 5, 453–455. [Google Scholar]
- Wagner, A.P.; Creel, S.; Kalinowski, S.T. Estimating relatedness and relationships using microsatellite loci with null alleles. Heredity 2006, 97, 336–345. [Google Scholar]
- Chapuis, M.-P.; Estoup, A. Microsatellite null alleles and estimation of population differentiation. Mol. Biol. Evol 2007, 24, 621–631. [Google Scholar]
- Lemer, S.; Rochel, E.; Planes, S. Correction method for null alleles in species with variable microsatellite flanking regions, a case study of the black-lipped pearl oysterPinctada margaritifera. J. Hered 2011, 102, 243–246. [Google Scholar]
- An, J.; Bechet, A.; Berggren, A.; Brwon, S.K.; Bruford, M.W.; Cai, Q.; Cassel-Lundhagen, A.; Cezilly, F.; Chen, S.L.; et al. Molecular Ecology Resources Primer Development Consortium. Permanent genetic resources added to molecular ecology resources database 1 October 2009–30 November 2009. Mol. Ecol. Resour 2010, 10, 404–408. [Google Scholar]
- Radosavljević, I.; Jakse, J.; Javornik, B.; Satovic, Z.; Liber, Z. New microsatellite markers for Salvia officinalis (Lamiaceae) and cross-amplification in closely related species. Am. J. Bot 2011, 98, e316–e318. [Google Scholar]
- Arnaud-Haond, S.; Belkhir, K. Genclone: A computer program to analyse genotypic data, test for clonality and describe spatial clonal organization. Mol. Ecol. Notes 2007, 7, 7–11. [Google Scholar]
- Arnaud-Haond, S.; Duarte, C.M.; Alberto, F.; Serrão, E.A. Standardizing methods to address clonality in population studies. Mol. Ecol 2007, 16, 5115–5139. [Google Scholar]
- Hamrick, J.L.; Godt, M.J.W. Allozyme Diversity in Plant Species. In Plant Population Genetics, Breeding and Genetic Resources; Brown, A.H.D., Clegg, M.T., Kahler, A.L., Weir, B.S., Eds.; Sinauer Associates Inc: Sunderland, MA, USA, 1989; pp. 43–63. [Google Scholar]
- Piñeiro, R.; Fuertes Aguilar, J.; Menezes de Sequeira, M.; Nieto Feliner, G. Low genetic diversity in the rare Madeiran endemic A. maderensis (Plumbaginaceae). Folia Geobot 2009, 44, 65–81. [Google Scholar]
- Ellegren, H.; Primmer, C.; Sheldon, B.C. Micro-satellite ‘evolution’: Directionality or bias. Nat. Genet 1995, 11, 360–362. [Google Scholar]
- Forbes, S.H.; Hogg, J.T.; Buchanan, E.C.; Crawford, A.M.; Allendorf, E.W. Microsatellite evolution in congeneric mammals: domestic and bighorn sheep. Mol. Biol. Evol 1995, 12, 1106–1113. [Google Scholar]
- Barker, J.S. Population structure and host-plant specialization in two Scaptodrosophila flower-breeding species. J. Hered 2005, 94, 129–138. [Google Scholar]
- Gitzendanner, M.A.; Soltis, P.S. Patterns of genetic variation in rare and widespread plant congeners. Am. J. Bot 2000, 87, 777–786. [Google Scholar]
- Felsenstein, J. Phylogenies and the comparative method. Am. Nat 1985, 125, 1–15. [Google Scholar]
- Cornuet, J.M.; Luikart, G. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 1996, 144, 1119–1127. [Google Scholar]
- IUCN Standards and Petitions Subcommittee. Guidelines for Using the IUCN Red List Categories and Criteria. Version 8.1. Prepared by the Standards and Petitions Subcommittee in March 2010. 2010. Available online: http://intranet.iucn.org/webfiles/doc/SSC/RedList/RedListGuidelines.pdf accessed on 26 June 2012.
- Jakše, J.; Javornik, B. High throughput isolation of microsatellites in hop (Humulus lupulus L.). Plant Mol. Biol. Rep 2001, 19, 217–226. [Google Scholar]
- Hamilton, M.B.; Pincus, E.L.; DiFiore, A.; Fleischer, R.C. Universal linker and ligation procedures for construction of genomic DNA libraries enriched for microsatellites. Biotechniques 1999, 27, 500–507. [Google Scholar]
- Thiel, T.; Michalek, W.; Varshney, R.K.; Graner, A. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor. Appl. Genet 2003, 106, 411–422. [Google Scholar]
- Rozen, S.; Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol 2000, 132, 365–386. [Google Scholar]
- Schuelke, M. An economic method for florescent labeling of PCR fragments. Nat. Biotechnol 2000, 18, 233–234. [Google Scholar]
- Botstein, D.; White, R.L.; Sholnick, M.; David, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet 1980, 32, 314–331. [Google Scholar]
- Liu, K.; Muse, S.V. PowerMarker—An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar]
- Raymond, M.; Rousset, F. GenePop (Version 1.2): Population genetics software for exact tests and ecumenicism. J. Hered 1995, 86, 248–249. [Google Scholar]
- Holm, S. A simple sequentially rejective multiple test procedure. Scand. J. Statist 1979, 6, 65–70. [Google Scholar]
- Van Oosterhout, C.; Hutchinson, W.F.; Wills, D.P.M.; Shipley, P. Micro-Checker: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar]
- Piry, S.; Luikart, G.; Cornuet, J.-M. Bottleneck: A computer programme for detecting recent reductions in the effective population size using allele frequency data. J. Hered 1999, 90, 502–503. [Google Scholar]
- Di Rienzo, A.; Peterson, A.C.; Garza, J.C.; Valdes, A.M.; Slatkin, M.; Freimer, N.B. Mutational process of simple-sequence repeat loci in human populations. Proc. Natl. Acad. Sci. USA 1994, 91, 3166–3170. [Google Scholar]
- Luikart, G.; Allendorf, F.W.; Cornuet, J.M.; Sherwin, W.B. Distortion of allele frequency distributions provides a test for recent population bottlenecks. J. Hered 1998, 89, 238–247. [Google Scholar]

| Locus name | Repeat motif | Primer sequence (5′–3′) | Size (bp) | Na | HO | HE | PIC | GenBank accession No. |
|---|---|---|---|---|---|---|---|---|
| SoUZ021 | (CA)13 | F: CATTCTTTGCAGGGATTCGT R: GATGCTTCCTCGGCTGACTA | 226–242 | 8 | 0.50 | 0.73 | 0.70 | JX440363 |
| SoUZ022 | (AG)19 | F: TCTTCGAGCCTGGAGTTTT R: AGAAGCAAGACAACCCCAAA | 226–264 | 18 | 0.54 | 0.89* | 0.89 | JX440364 |
| SoUZ023 | (AC)14 | F: CCTGCAAAACACAAACGAA R: GTTGTTTCGCTGGTGATGAA | 171–185 | 8 | 0.83 | 0.83 | 0.81 | JX440365 |
| SoUZ024 | (GA)25 | F: TGGTCGTGTTGAACTTTCG R: AAGGAAGGTGCACCAAAATG | 128–177 | 18 | 0.65 | 0.89 | 0.89 | JX440366 |
| SoUZ025 | (AG)31 | F: AGGTGTGTGACCCTGCTATG R: GGTTTTGCTCCATTGCATTT | 205–246 | 21 | 0.67 | 0.93* | 0.92 | JX440367 |
| SoUZ026 | (AG)17 | F: TTCATCTTTGACCGGAAAAC R: CATGTGGTGATGCGAGATTC | 160–191 | 13 | 0.67 | 0.86 | 0.85 | JX440368 |
| SoUZ027 | (AG)24 | F: GGCGAGATTCATTTCCTTGA R: CATCAGTGAGGCTTGGTTCA | 196–240 | 14 | 0.46 | 0.84* | 0.83 | JX440369 |
| SoUZ028 | (AG)19 | F: GGGCCTTGTCTGCATGTATT R: TCCGGCGATTGTTCTCTAAT | 201–235 | 15 | 0.71 | 0.88 | 0.87 | JX440370 |
| SoUZ029 | (GT)13 | F: AAACACGCATTTGTACGTGAA R: CCAACGACAACATCATCGTC | 155–182 | 10 | 0.52 | 0.84 | 0.81 | JX440371 |
| Locus | Salvia officinalis (N = 25) | Salvia brachyodon (N = 25) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Size (bp) | Na | HO | HE | PIC | Size (bp) | Na | HO | HE | PIC | CA | |
| SoUZ001a | 159–195 | 15 | 0.83 | 0.90 | 0.90 | 158–172 | 4 | 0.72 | 0.66 | 0.62 | - |
| SoUZ002a | 199–222 | 11 | 0.60 | 0.77 | 0.74 | 190–194 | 3 | 0.67 | 0.54 | 0.47 | - |
| SoUZ004a | 195–227 | 12 | 0.62 | 0.82 | 0.80 | 199–212 | 6 | 0.58 | 0.60 | 0.57 | 2 |
| SoUZ005a | 131–160 | 9 | 0.30 | 0.81* | 0.79 | 122–130 | 5 | 0.83 | 0.77 | 0.73 | - |
| SoUZ006a | 204–244 | 14 | 0.83 | 0.88 | 0.87 | 204–233 | 9 | 0.76 | 0.84 | 0.82 | 6 |
| SoUZ007a | 214–227 | 8 | 0.58 | 0.72 | 0.69 | 210–240 | 9 | 0.92 | 0.84 | 0.82 | 5 |
| SoUZ008a | 190–210 | 10 | 0.71 | 0.80 | 0.77 | 176–180 | 3 | 0.80 | 0.65 | 0.57 | - |
| SoUZ009a | 209–250 | 9 | 0.61 | 0.74 | 0.70 | 220–235 | 6 | 0.72 | 0.73 | 0.70 | 2 |
| SoUZ011a | 175–227 | 19 | 0.76 | 0.93 | 0.93 | 230–250 | 6 | 0.92 | 0.77 | 0.74 | - |
| SoUZ014b | 201–237 | 12 | 0.87 | 0.88 | 0.87 | 192–218 | 9 | 0.75 | 0.82 | 0.80 | 2 |
| SoUZ020b | 198–246 | 10 | 0.50 | 0.77 | 0.74 | 199–217 | 4 | 0.33 | 0.62* | 0.55 | - |
| SoUZ021 | 226–242 | 8 | 0.50 | 0.73 | 0.70 | 224–238 | 5 | 0.58 | 0.69 | 0.65 | 4 |
| SoUZ023 | 171–185 | 8 | 0.83 | 0.83 | 0.81 | 167–183 | 6 | 0.75 | 0.76 | 0.72 | 2 |
| SoUZ024 | 128–177 | 18 | 0.65 | 0.89 | 0.89 | 116–134 | 8 | 0.83 | 0.75 | 0.72 | 2 |
| SoUZ026 | 160–191 | 13 | 0.67 | 0.86 | 0.85 | 166–176 | 4 | 0.37 | 0.33 | 0.31 | 4 |
© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Radosavljević, I.; Satovic, Z.; Jakse, J.; Javornik, B.; Greguraš, D.; Jug-Dujaković, M.; Liber, Z. Development of New Microsatellite Markers for Salvia officinalis L. and Its Potential Use in Conservation-Genetic Studies of Narrow Endemic Salvia brachyodon Vandas. Int. J. Mol. Sci. 2012, 13, 12082-12093. https://doi.org/10.3390/ijms130912082
Radosavljević I, Satovic Z, Jakse J, Javornik B, Greguraš D, Jug-Dujaković M, Liber Z. Development of New Microsatellite Markers for Salvia officinalis L. and Its Potential Use in Conservation-Genetic Studies of Narrow Endemic Salvia brachyodon Vandas. International Journal of Molecular Sciences. 2012; 13(9):12082-12093. https://doi.org/10.3390/ijms130912082
Chicago/Turabian StyleRadosavljević, Ivan, Zlatko Satovic, Jernej Jakse, Branka Javornik, Danijela Greguraš, Marija Jug-Dujaković, and Zlatko Liber. 2012. "Development of New Microsatellite Markers for Salvia officinalis L. and Its Potential Use in Conservation-Genetic Studies of Narrow Endemic Salvia brachyodon Vandas" International Journal of Molecular Sciences 13, no. 9: 12082-12093. https://doi.org/10.3390/ijms130912082
APA StyleRadosavljević, I., Satovic, Z., Jakse, J., Javornik, B., Greguraš, D., Jug-Dujaković, M., & Liber, Z. (2012). Development of New Microsatellite Markers for Salvia officinalis L. and Its Potential Use in Conservation-Genetic Studies of Narrow Endemic Salvia brachyodon Vandas. International Journal of Molecular Sciences, 13(9), 12082-12093. https://doi.org/10.3390/ijms130912082







