Evaluation of HER2 Gene Amplification in Breast Cancer Using Nuclei Microarray in Situ Hybridization
Abstract
:1. Introduction
2. Results and Discussion
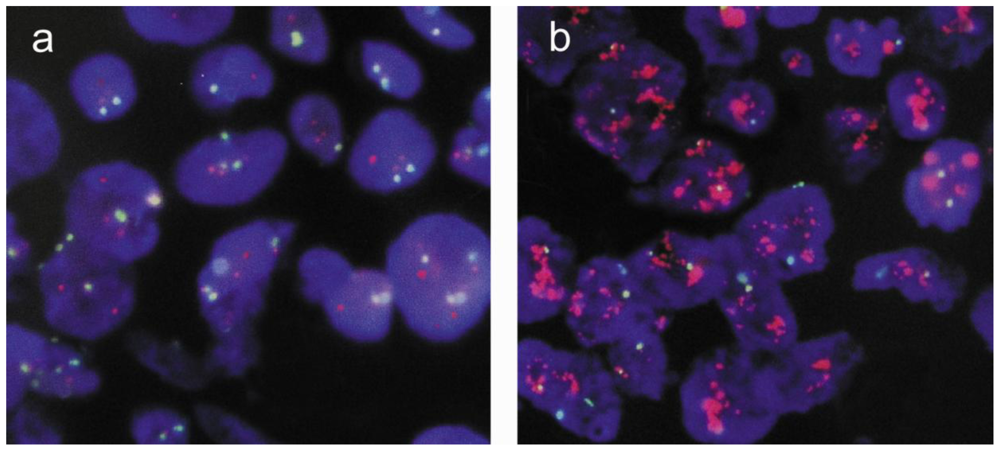
2.1. Results
2.2. Discussion
3. Experimental Section
3.1. Nuclei Microarray Construction
3.2. Nuclei Microarrays Fluorescence in Situ Hybridization (NMFISH)
3.3. Conventional Fluorescence in Situ Hybridization (FISH)
3.4. Evaluation of HER2 Gene Amplification
3.5. Statistics
4. Conclusions
Acknowledgments
- Conflict of InterestThere is no potential or actual personal, financial or political interest related to this article.
References
- Gschwind, A.; Fischer, O.M.; Ullrich, A. The discovery of receptor tyrosine kinases: Targets for cancer therapy. Nat. Rev. Cancer 2004, 4, 361–370. [Google Scholar]
- Jones, F.E.; Stern, D.F. Expression of dominant-negative ErbB2 in the mammary gland of transgenic mice reveals a role in lobuloalveolar development and lactation. Oncogene 1999, 18, 3481–3490. [Google Scholar]
- Slamon, D.J.; Godolphin, W.; Jones, L.A.; Holt, J.A.; Wong, S.G.; Keith, D.E.; Levin, W.J.; Stuart, S.G.; Udove, J.; Ullrich, A.; et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science 1989, 244, 707–712. [Google Scholar]
- Cobleigh, M.A.; Vogel, C.L.; Tripathy, D.; Robert, N.J.; Scholl, S.; Fehrenbacher, L.; Wolter, J.M.; Paton, V.; Shak, S.; Lieberman, G.; et al. Multinational study of the efficacy and safety of humanized anti-HER2 monoclonal antibody in women who have HER2-overexpressing metastatic breast cancer that has progressed after chemotherapy for metastatic disease. J. Clin. Oncol 1999, 17, 2639–2648. [Google Scholar]
- Cai, F.F.; Kohler, C.; Zhang, B.; Wang, M.H.; Chen, W.J.; Zhong, X.Y. Epigenetic therapy for breast cancer. Int. J. Mol. Sci 2011, 12, 4465–4487. [Google Scholar]
- Pritchard, K.I.; Shepherd, L.E.; O’Malley, F.P.; Andrulis, I.L.; Tu, D.; Bramwell, V.H.; Levine, M.N. HER2 and responsiveness of breast cancer to adjuvant chemotherapy. N. Engl. J. Med 2006, 354, 2103–2111. [Google Scholar]
- Arteaga, C.L.; Sliwkowski, M.X.; Osborne, C.K.; Perez, E.A.; Puglisi, F.; Gianni, L. Treatment of HER2-positive breast cancer: Current status and future perspectives. Nat. Rev. Clin. Oncol 2012, 9, 16–32. [Google Scholar]
- Baselga, J.; Bradbury, I.; Eidtmann, H.; Di, C.S.; de Azambuja, E.; Aura, C.; Gomez, H.; Dinh, P.; Fauria, K.; van Dooren, V.; et al. Lapatinib with trastuzumab for HER2-positive early breast cancer (NeoALTTO): A randomised, open-label, multicentre, phase 3 trial. Lancet 2012, 379, 633–640. [Google Scholar]
- Zhang, W.; Yu, Y. The important molecular markers on chromosome 17 and their clinical impact in breast cancer. Int. J. Mol. Sci 2011, 12, 5672–5683. [Google Scholar]
- Slodkowska, J.; Filas, V.; Buszkiewicz, E.; Trzeciak, P.; Wojciechowski, M.; Koktysz, R.; Staniszewski, W.; Breborowicz, J.; Rojo, M.G. Study on breast carcinoma HER2/neu and hormonal receptors status assessed by automated images analysis systems: ACIS III (Dako) and ScanScope (Aperio). Folia Histochem. Cytobiol 2010, 48, 19–25. [Google Scholar]
- Wolff, A.C.; Hammond, M.E.; Schwartz, J.N.; Hagerty, K.L.; Allred, D.C.; Cote, R.J.; Dowsett, M.; Fitzgibbons, P.L.; Hanna, W.M.; Langer, A.; et al. American society of clinical oncology/college of American pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J. Clin. Oncol 2007, 25, 118–145. [Google Scholar]
- Shah, S.S.; Wang, Y.; Tull, J.; Zhang, S. Effect of high copy number of HER2 associated with polysomy 17 on HER2 protein expression in invasive breast carcinoma. Diagn. Mol. Pathol 2009, 18, 30–33. [Google Scholar]
- Pedersen, M.; Rasmussen, B.B. The correlation between dual-color chromogenic in situ hybridization and fluorescence in situ hybridization in assessing HER2 gene amplification in breast cancer. Diagn. Mol. Pathol 2009, 18, 96–102. [Google Scholar]
- Meijer, S.L.; Wesseling, J.; Smit, V.T.; Nederlof, P.M.; Hooijer, G.K.; Ruijter, H.; Arends, J.W.; Kliffen, M.; van, G.J.M.; Sterk, L.; et al. HER2 gene amplification in patients with breast cancer with equivocal IHC results. J. Clin. Pathol 2011, 64, 1069–1172. [Google Scholar]
- Tanner, M.; Gancberg, D.; Di, L.A.; Larsimont, D.; Rouas, G.; Piccart, M.J.; Isola, J. Chromogenic in situ hybridization: A practical alternative for fluorescence in situ hybridization to detect HER-2/neu oncogene amplification in archival breast cancer samples. Am. J. Pathol 2000, 157, 1467–1472. [Google Scholar]
- Rimm, D.L.; Camp, R.L.; Charette, L.A.; Olsen, D.A.; Provost, E. Amplification of tissue by construction of tissue microarrays. Exp. Mol. Pathol 2001, 70, 255–264. [Google Scholar]
- Moskaluk, C.A.; Stoler, M.H. Agarose mold embedding of cultured cells for tissue microarrays. Diagn. Mol. Pathol 2002, 11, 234–238. [Google Scholar]
- Jiang, H.Y.; Zhang, S.Q.; Zhao, T. A new method to make nuclei or cell microarrays. Diagn. Mol. Pathol 2006, 15, 109–114. [Google Scholar]
- Kononen, J.; Bubendorf, L.; Kallioniemi, A.; Barlund, M.; Schraml, P.; Leighton, S.; Torhorst, J.; Mihatsch, M.J.; Sauter, G.; Kallioniemi, O.P. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat. Med 1998, 4, 844–847. [Google Scholar]
- Von Wasielewski, R.; Mengel, M.; Wiese, B.; Rudiger, T.; Muller-Hermelink, H.K.; Kreipe, H. Tissue array technology for testing interlaboratory and interobserver reproducibility of immunohistochemical estrogen receptor analysis in a large multicenter trial. Am. J. Clin. Pathol 2002, 118, 675–682. [Google Scholar]
- Braunschweig, T.; Chung, J.Y.; Hewitt, S.M. Tissue microarrays: Bridging the gap between research and the clinic. Expert Rev. Proteomics 2005, 2, 325–336. [Google Scholar]
- Paternoster, S.F.; Brockman, S.R.; McClure, R.F.; Remstein, E.D.; Kurtin, P.J.; Dewald, G.W. A new method to extract nuclei from paraffin-embedded tissue to study lymphomas using interphase fluorescence in situ hybridization. Am. J. Pathol 2002, 160, 1967–1972. [Google Scholar]
- Landis, J.R.; Koch, G.G. A one-way components of variance model for categorical data. Biometrics 1977, 334, 671–679. [Google Scholar]
| Nuclei Microarray FISH | ||||
|---|---|---|---|---|
| FISH | Non-Amplified * | Equivocal § | Amplified ‡ | Total |
| Non-amplified * | 84 | 3 | 0 | 87 |
| Equivocal § | 2 | 19 | 2 | 23 |
| Amplified ‡ | 0 | 0 | 42 | 42 |
| Total | 86 | 22 | 44 | 152 |
| Case # | NMFISH/FISH Status | NMFISH | FISH |
|---|---|---|---|
| 1# | Non-amplified/equivocal | 1.74 | 1.86 |
| 2# | Non-amplified/equivocal | 1.67 | 1.85 |
| 3# | Equivocal/Non-amplified | 1.87 | 1.71 |
| 4# | Equivocal/Non-amplified | 1.91 | 1.65 |
| 5# | Equivocal/Non-amplified | 1.86 | 1.74 |
| 6# | Amplified/equivocal | 2.39 | 2.15 |
| 7# | Amplified/equivocal | 2.45 | 2.13 |
© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Jiang, H.; Bai, X.; Zhang, C.; Zhang, X. Evaluation of HER2 Gene Amplification in Breast Cancer Using Nuclei Microarray in Situ Hybridization. Int. J. Mol. Sci. 2012, 13, 5519-5527. https://doi.org/10.3390/ijms13055519
Jiang H, Bai X, Zhang C, Zhang X. Evaluation of HER2 Gene Amplification in Breast Cancer Using Nuclei Microarray in Situ Hybridization. International Journal of Molecular Sciences. 2012; 13(5):5519-5527. https://doi.org/10.3390/ijms13055519
Chicago/Turabian StyleJiang, Huiyong, Xiaoyan Bai, Cheng Zhang, and Xuefeng Zhang. 2012. "Evaluation of HER2 Gene Amplification in Breast Cancer Using Nuclei Microarray in Situ Hybridization" International Journal of Molecular Sciences 13, no. 5: 5519-5527. https://doi.org/10.3390/ijms13055519




