Heat Shock Protein 90 in Plants: Molecular Mechanisms and Roles in Stress Responses
Abstract
:1. Introduction
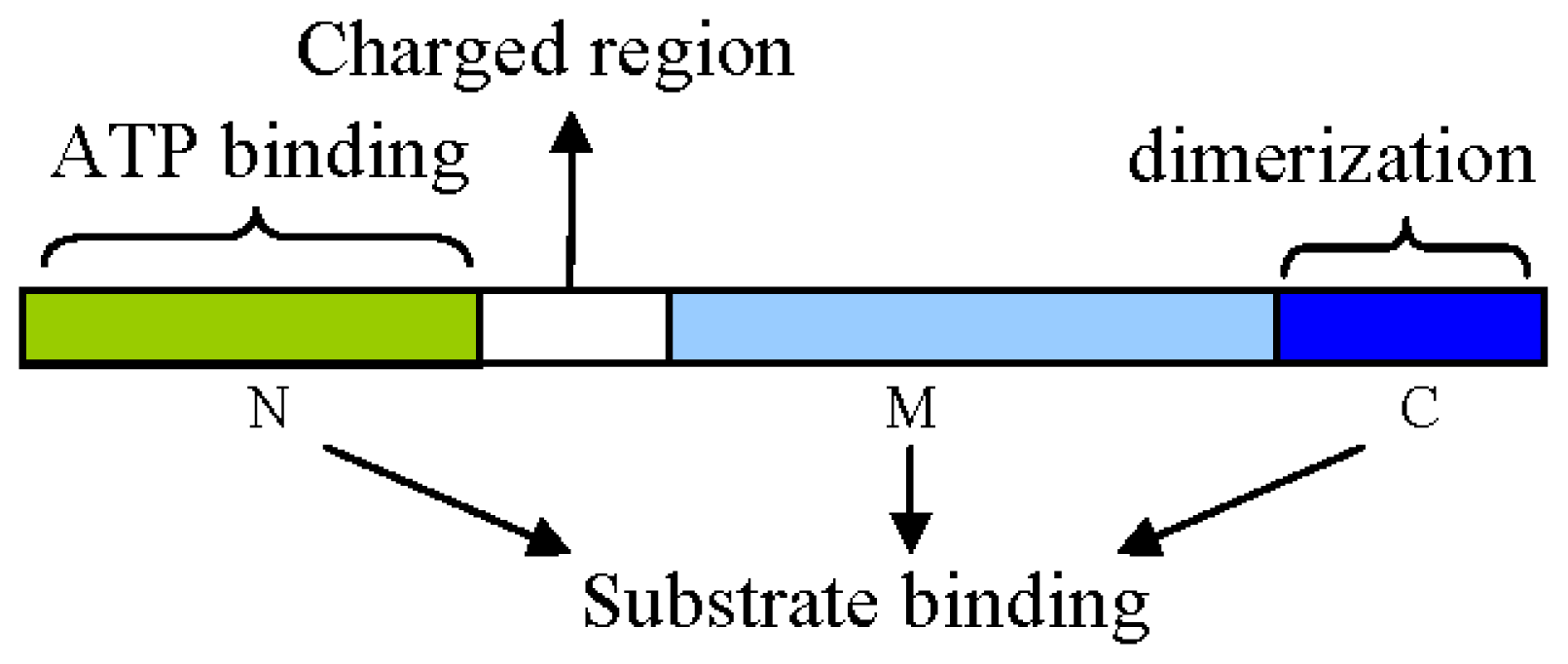
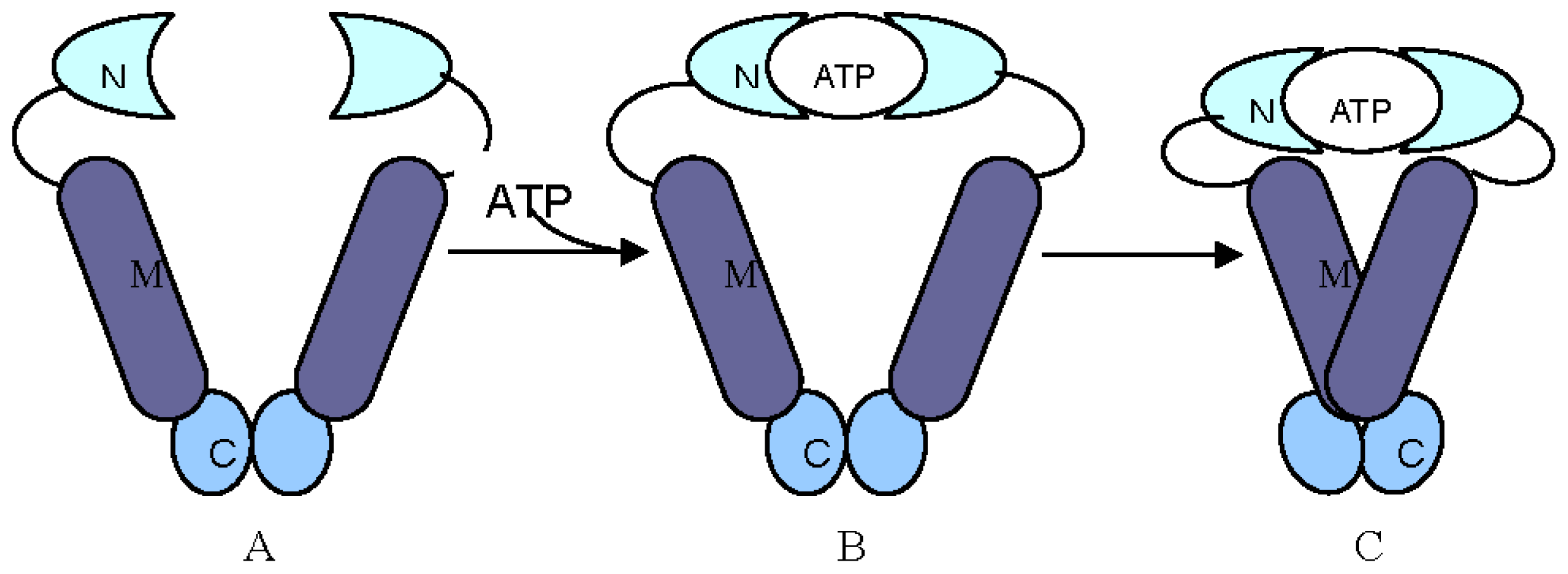
2. Structural and Functional Analyses of Hsp90
3. Interactions of Proteins with Hsp90s and Their Interaction Mechanism
3.1. Accessory Proteins
3.2. Regulators
3.3. Substrate Proteins
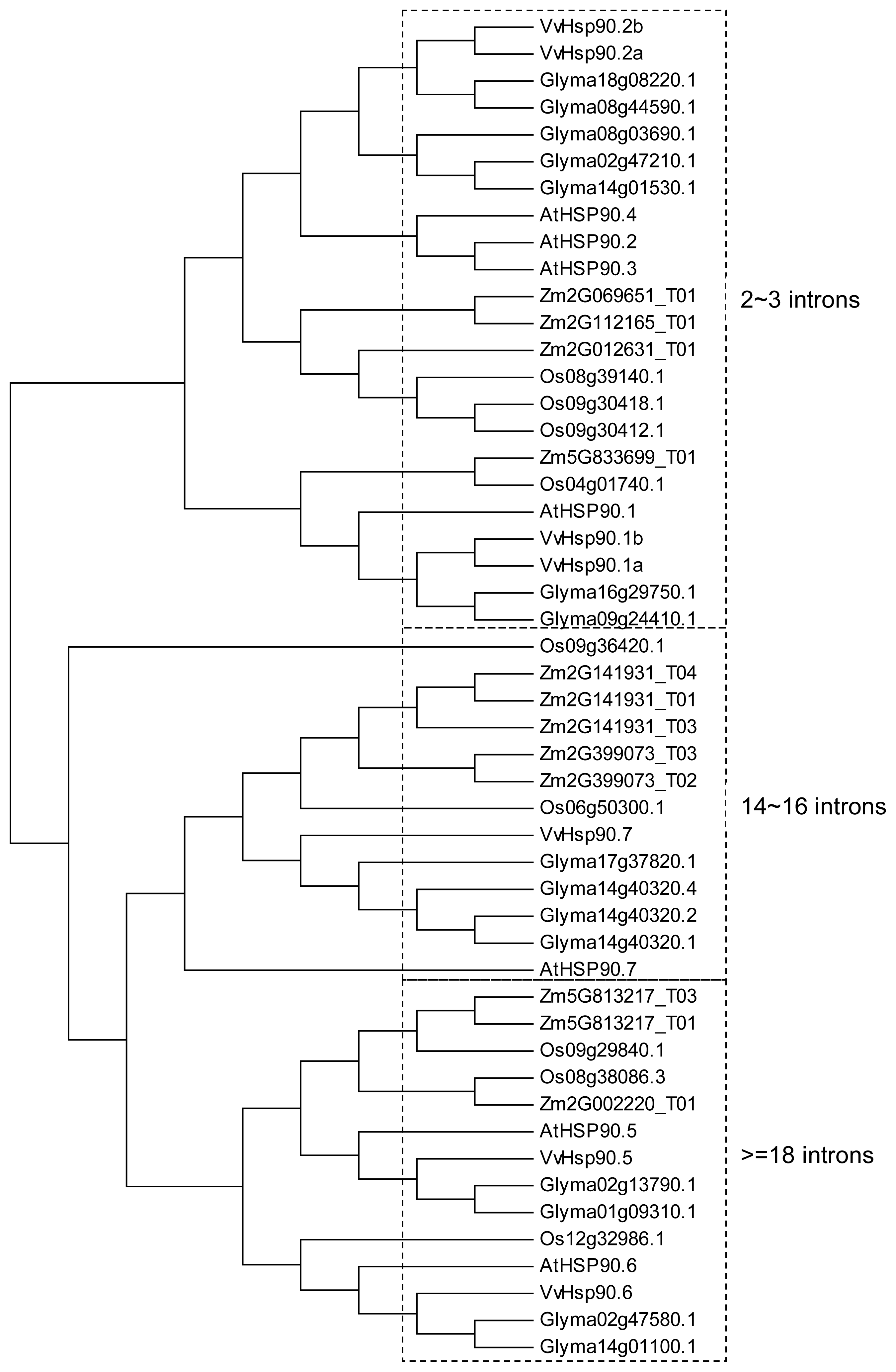
4. The Hsp90 Family in Model Plants
5. Hsp90 Is Involved in Plant Stress Resistance
5.1. Hsp90s Are Involved in Disease and Pest Resistances
5.2. Hsp90s Mediate Abiotic Stress Resistance
6. Prospects
Acknowledgments
References
- Lindquist, S. The heat-shock response. Annu. Rev. Biochem 1986, 55, 1151–1191. [Google Scholar]
- Lindquist, S.; Craig, E.A. The heat-shock proteins. Annu. Rev. Genetics 1988, 22, 631–677. [Google Scholar]
- Horwitz, J. Alpha-crystallin can function as a molecular chaperone. Proc. Natl. Acad. Sci. USA 1992, 89, 10449–10453. [Google Scholar]
- Picard, D. Heat-shock protein 90, a chaperone for folding and regulation. Cell. Mol. Life Sci 2002, 59, 1640–1648. [Google Scholar]
- Jackson, S.E.; Queistch, C.; Toft, D. Hsp90: From structure to phenotype. Nat. Struct. Mol. Biol 2004, 11, 1152–1155. [Google Scholar]
- Wegele, H.; Muller, L.; Buchner, J. Hsp70 and Hsp90-a relay team for protein folding. Rev. Physiol. Biochem. Pharmacol 2004, 151, 1–44. [Google Scholar]
- Shinozaki, F.; Minami, M.; Chiba, T.; Suzuki, M.; Yoshimatsu, K.; Ichikawa, Y.; Terasawa, K.; Emori, Y.; Matsumoto, K.; Kurosaki, T.; et al. Depletion of Hsp90β induces multiple defects in B cell receptor signaling. J. Biol. Chem 2006, 281, 16361–16369. [Google Scholar]
- Zuehlke, A.; Johnson, J.L. Hsp90 and co-chaperones twist the functions of diverse client proteins. Biopolymers 2010, 93, 211–217. [Google Scholar]
- Scroggins, B.T.; Robzyk, K.; Wang, D.; Marcu, M.G.; Tsutsumi, S.; Beebe, K.; Cotter, R.J.; Felts, S.; Toft, D.; Karnitz, L.; et al. An acetylation site in the middle domain of Hsp90 regulates chaperone function. Mol. Cell 2007, 25, 151–159. [Google Scholar]
- Kamal, A.; Thao, L.; Sensintaffar, J.; Zhang, L.; Boehm, M.F.; Fritz, L.C.; Burrows, F.J. A high-affinity conformation of Hsp90 confers tumour selectivity on Hsp90 inhibitors. Nature 2003, 425, 407–410. [Google Scholar]
- Xu, W.; Neckers, L. Targeting the molecular chaperone Heat Shock Protein 90 provides a multifaceted effect on diverse cell signaling pathways of cancer cells. Clin. Cancer Res 2007, 13, 1625–1629. [Google Scholar]
- Ali, M.M.; Roe, S.M.; Vaughan, C.K.; Meyer, P.; Panaretou, B.; Piper, P.W.; Prodromou, C.; Pearl, L.H. Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature 2006, 440, 1013–1017. [Google Scholar]
- Chen, Z.; Sasaki, T.; Tan, X.; Carretero, J.; Shimamura, T.; Li, D.; Xu, C.; Wang, Y.; Aldelmant, G.O.; Capelletti, M.; et al. Inhibition of ALK, PI3K/MEK and HSP90 in murine lung adenocarcinoma induced by EML4-ALK fusion oncogene. Cancer Res 2010, 70, 9827–9836. [Google Scholar]
- Grigorova, B.; Vaseva, I.; Demirevska, K.; Feller, U. Combined drought and heat stress in wheat: Changes in some heat shock proteins. Biol. Plant 2011, 55, 105–111. [Google Scholar]
- Maksymied, W. Effects of jasmonates and some other signaling factors on bean and onion growth during the initial phase of cadmium action. Biol. Plant 2011, 55, 112–118. [Google Scholar]
- Wójcik, M.; Tukiendorf, A. Glutathione in adaptation of Arabidopsis thaliana to cadmium stress. Biol. Plant 2011, 55, 125–132. [Google Scholar]
- Zhang, L.; Fan, Y.; Shi, F.; Qin, S.; Liu, B. Moleculat cloning, characterization, and expression analysis of a cytosolic HSP90 gene from Haematococcus pluvialis. J. Appl. Phycol. 2012. [Google Scholar] [CrossRef]
- Banilas, G.; Korkas, E.; Englezos, V.; Nisiotou, A.A.; Hatzopoulos, P. Genome-wide analysis of the heat shock protein 90 gene family in grapevine (Vitis vinifera L.). Aust. J. Grape Wine Res 2012, 18, 29–38. [Google Scholar]
- Rizhsky, L.; Liang, H.; Mittler, R. The combined effect of drought stress and heat shock on gene expression in tobacco. Plant Physiol 2002, 130, 1143–1151. [Google Scholar]
- Sangster, T.A.; Queitsch, C. The HSP90 chaperone complex, an emerging force in plant development and phenotypic plasticity. Curr. Opin. Plant Biol 2005, 8, 86–92. [Google Scholar]
- Lindquist, S.; Jarosz, D.F. Hsp90 and environmental stress transform the adaptive value of natural genetic variation. Science 2010, 330, 1820–1824. [Google Scholar]
- Sangster, T.A.; Salathia, N.; Lee, H.N.; Watanabe, E.; Schellenberg, K.; Morneau, K.; Wang, H.; Undurraga, S.; Queitsch, C.; Lindquist, S. HSP90-buffered genetic variation is common in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2008, 105, 2969–2974. [Google Scholar]
- Young, J.C.; Moarefi, I.; Hartl, F.U. Hsp90: A specialized but essential protein-folding tool. J. Cell Biol 2001, 154, 267–273. [Google Scholar]
- Gupta, R.S. Phylogenetic analysis of the 90kD heat shock family of protein sequences and an examination of the relationship among animals, plant, and fungi species. Mol. Biol. Evol 1995, 12, 1063–1073. [Google Scholar]
- Hao, H.; Naomoto, Y.; Bao, X.; Watanabe, N.; Sakurama, K.; Noma, K.; Motoki, T.; Tomono, Y.; Fukazawa, T.; Shirakawa, Y.; et al. HSP90 and its inhibitors. Oncol. Rep 2010, 23, 1483–1492. [Google Scholar]
- Ogiso, H.; Kagi, N.; Matsumoto, E.; Nishimoto, M.; Arai, R.; Shirouzu, M.; Mimura, J.; Fujii-Kuriyama, Y.; Yokoyama, S. Phosphorylation analysis of 90 kDa heat shock protein within the cytosolic arylhydrocarbon receptor complex. Biochemistry 2004, 43, 15510–15519. [Google Scholar]
- Parsell, D.A.; Lindquist, S. The function of heat-shock proteins in stress tolerance: Degradation and reactivation of damaged proteins. Annu. Rev. Genet 1993, 27, 437–496. [Google Scholar]
- Csermely, P.; Schnaider, T.; Soti, C.; Prohaszka, Z.; Nardai, G. The 90-kDa molecular chaperone family: Structure, function, and clinical applications. A comprehensive review. Pharmacol. Ther 1998, 79, 129–168. [Google Scholar]
- Terasawa, K.; Minami, M.; Minami, Y. Constantly updated knowledge of Hsp90. J. Biochem 2005, 137, 443–447. [Google Scholar]
- Cowen, L.E. The evolution of fungal drug resistance: Modulating the trajectory from genotype to phenotype. Nat. Rev. Microbiol 2008, 6, 187–198. [Google Scholar]
- Wayne, N.; Mishra, P.; Bolon, D.N. Hsp90 and client protein maturation. Methods Mol. Biol 2011, 787, 33–44. [Google Scholar]
- Hessling, M.; Richter, K.; Buchener, J. Dissection of the ATP-induced conformational cycle of the molecular chaperone Hsp90. Nat. Struct. Mol. Biol 2009, 16, 287–293. [Google Scholar]
- Mickler, M.; Hessling, M.; Ratzke, C.; Buchner, J.; Hugel, T. The large conformational changes of Hsp90 are only weakly coupled to ATP hydrolysis. Nat. Struct. Mol. Boil 2009, 16, 281–286. [Google Scholar]
- Wegele, H.; Wandinger, S.K.; Schmid, A.B.; Reinstein, J.; Buchner, J. Substrate transfer from the chaperone Hsp70 to Hsp90. J. Mol. Biol 2006, 356, 802–811. [Google Scholar]
- Whitesell, L.; Shifrin, S.D.; Schwab, G.; Neckers, L.M. Benzoquinonoid ansamycins possess selective tumoricidal activity unrelated to src kinase inhibition. Cancer Res 1992, 52, 1721–1728. [Google Scholar]
- Weikl, T.; Muschler, P.; Richter, K.; Veit, T.; Reinstein, J.; Buchner, J. C-terminal regions of Hsp90 are important for trapping the nucleotide during the ATPase cycle. J. Mol. Biol 2000, 303, 583–592. [Google Scholar]
- Meyer, P.; Prodromou, C.; Hu, B.; Vaughan, C.; Roe, S.M.; Panaretou, B.; Piper, P.W.; Pearl, L.H. Structural and functional analysis of the middle segment of Hsp90: Implications for ATP hydrolysis and client protein and cochaperone interactions. Mol. Cell 2003, 11, 647–658. [Google Scholar]
- Pearl, L.H.; Prodromou, C. Structure and in vivo function of Hsp90. Curr. Opin. Struct. Biol 2000, 10, 46–51. [Google Scholar]
- Zhao, R.; Davey, M.; Hsu, Y.C.; Kaplanek, P.; Tong, A.; Parsons, A.B.; Krogan, N.; Cagney, G.; Mai, D.; Greenblatt, J.; et al. Navigating the chaperone network: An integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell 2005, 120, 715–727. [Google Scholar]
- Johnson, J.L.; Brown, C. Plasticity of the Hsp90 chaperone machine in divergent eukaryotic organisms. Cell Stress Chaperon 2009, 14, 83–94. [Google Scholar]
- Aviezer-Hagai, K.; Skovorodnikova, J.; Galigniana, M.; Farchi-Pisanty, O.; Maayan, E.; Bocovza, S.; Efrat, Y.; von Koskull-Döring, P.; Ohad, N.; Breiman, A. Arabidopsis immunophilins ROF1 (AtFKBP62) and ROF2 (AtFKBP65) exhibit tissue specificity, are heat-stress induced, and bind HSP90. Plant Mol. Biol. 2007, 63, 237–255. [Google Scholar]
- Weaver, A.J.; Sullivan, W.P.; Felts, S.J.; Owen, B.A.; Toft, D.O. Crystal structure and activity of human p23, a heat shock protein 90 co-chaperone. J. Mol. Biol 2000, 275, 23045–23052. [Google Scholar]
- Tinel, A.; Eckert, M.J.; Logette, E.; Lippens, S.; Janssens, S.; Jaccard, B.; Quadroni, M.; Tschopp, J. Regulation of PIDD auto-proteolysis and activity by the molecular chaperone Hsp90. Cell Death Differ 2010, 18, 506–515. [Google Scholar]
- Janssens, S.; Tinel, A.; Lippens, S.; Tschopp, J. PIDD mediates NF-κB activation in response to DNA damage. Cell 2005, 123, 1079–1092. [Google Scholar]
- Hunter, T.; Poon, R.Y.C. Cdc37: A protein kinase chaperone. Trends Cell. Biol 1997, 7, 157–161. [Google Scholar]
- Dmochewitz, L.; Lillich, M.; Kaiser, E.; Jennings, L.D.; Lang, A.E.; Buchner, J.; Fischer, G.; Aktories, K.; Collier, R.J.; Barth, H. Role of CypA and Hsp90 in membrane translocation mediated by anthrax protective antigen. Cell. Microbiol 2010, 13, 359–373. [Google Scholar]
- Szyszka, R.; Kramer, G.; Hardesty, B. The phosphorylation state of the reticulocyte 90-kDa heat shock protein affects its ability to increase phosphorylation of peptide initiation factor 2a subunit by the heme-sensitive kinase. Biochemistry 1989, 28, 1435–1438. [Google Scholar]
- Mimnaugh, E.G.; Worland, P.J.; Whitesell, L.; Neckers, L.M. Possible role for serine/threonine phosphorylation in the regulation of the heteroprotein complex between the hsp90 stress protein and the pp60v-src tyrosine kinase. J. Biol. Chem 1995, 270, 28654–28659. [Google Scholar]
- Morano, K.A.; Thiele, D.J. The Sch9 protein kinase regulates Hsp90 chaperone complex signal transduction activity in vivo. EMBO J 1999, 18, 5953–5962. [Google Scholar]
- Trott, A.; Shaner, L.; Morano, K.A. The molecular chaperone Sse1 and the growth control protein kinase Sch9 collaborate to regulate protein kinase A activity in Saccharomyces cerevisiae. Genetics 2005, 107, 1009–1021. [Google Scholar]
- Echeverria, P.C.; Flrafonov, F.; Pandey, D.P.; Muhlebach, G.; Picard, D. Detection of changes in gene regulatory patterns, elicited by perturbations of the Hsp90 molecular chaperone complex, by visualizing multiple experiments with an animation. BioData Min 2011, 4, 15. [Google Scholar]
- Flom, G.A.; Langner, E.; Johnson, J.L. Identification of an Hsp90 mutation that selectively disrupts cAMP/PKA signaling inSaccharomyces cerevisiae. Curr. Genet 2012. [Google Scholar] [CrossRef]
- Pratt, W.B.; Galigniana, M.D.; Harrell, J.M.; DeFranco, D.B. Role of hsp90 and the hsp90-binding immunophilins in signalling protein movement. Cell Signal 2004, 16, 857–872. [Google Scholar]
- Krukenberg, K.A.; Street, T.O.; Lavery, L.A.; Agard, D.A. Conformational dynamics of the molecular chaperone Hsp90. Q. Rev. Biophys 2011, 44, 229–255. [Google Scholar]
- Milioni, D.; Hatzopoulos, P. Genonic organization of hsp90 gene family in Arabidopsis. Plant Mol. Biol. 1997, 35, 955–961. [Google Scholar]
- Sato, S.; Fujita, N.; Tsuruo, T. Modulation of Akt kinase activity by binding to Hsp90. Proc. Natl. Acad. Sci 2000, 97, 10832–10837. [Google Scholar]
- Bijlmakers, M.J.; Marsh, M. Hsp90 is essential for the synthesis and subsequent membrane association, but not the maintenance, of the Src-kinase p56lck. Mol. Biol. Cell 2000, 11, 1585–1595. [Google Scholar]
- Lee, J.H.; Chung, I.K. Curcumin inhibits nuclear localization of telomerase by dissociating the Hsp90 co-chaperone p23 from hTERT. Cancer Lett 2010, 290, 76–86. [Google Scholar]
- Street, T.O.; Lavery, L.A.; Agard, D.A. Substrate binding drives large-scale conformational changes in the Hsp90 molecular chaperone. Mol. Cell 2011, 42, 96–105. [Google Scholar]
- Samakovli, D.; Thanou, A.; Hztzopoulos, P. Hsp90 canalizes developmental perturbation. J. Exp. Bot 2007, 58, 3513–3524. [Google Scholar]
- Specchia, V.; Piacentini, L.; Tritto, P.; Fanti, L.; D’Alessandro, R.; Palumbo, G.; Pimpinelli, S.; Bozzetti, M.P. Hsp90 prevents phenotypic variation by suppressing the mutagenic acivity of transposons. Nature 2010, 463, 662–665. [Google Scholar]
- TAIR. Available online: http://www.arabidopsis.org accessed on 13 November 2012.
- Plant Biology. Available online: http://www.rice.plantbiology.msu.edu accessed on 13 November 2012.
- Phytozome. Available online: http://www.phytozome.net/soybean accessed on 13 November 2012.
- MaizeGDB. Available online: http://www.maizegdb.org accessed on 13 November 2012.
- Krishna, P.; Gloor, G. The Hsp90 family of proteins in Arabidopsis thaliana. Cell Stress Chaperon 2001, 6, 238–246. [Google Scholar]
- PSORT. Available online: http://psort.nibb.ac.jp/form.html accessed on 13 November 2012.
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple sequence alignment using ClustalW and ClustalX. In Curr. Protoc. Bioinforma; 2002; pp. 2.3.1–2.3.22. [Google Scholar]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol 2007, 24, 1596–1599. [Google Scholar]
- Queitsch, C.; Sangster, T.A.; Lindquist, S. Hsp90 as a capacitor of phenotypic variation. Nature 2002, 417, 598–599. [Google Scholar]
- Sangster, T.A.; Bahrami, A.; Wilczek, A.; Watanabe, E.; Schellenberg, K.; McLellan, C.; Kelley, A.; Kong, S.W.; Queitsch, C.; Lindquist, S. Phenotypic diversity and altered environmental plasticity in Arabidopsis thaliana with reduced Hsp90 levels. PLoS One 2007, 2, e648. [Google Scholar]
- Prasinos, C.; Krampis, K.; Samakovli, D.; Hatzopoulos, P. Tight regulation of expression of two Arabidopsis cytosolic HSP90 genes during embryo development. J. Exp. Bot 2005, 56, 633–644. [Google Scholar]
- Sangster, T.A.; Salathia, N.; Undurraga, S.; Milo, R.; Schellenberg, K.; Lindquist, S.; Queitsch, C. HSP90 affects the expression of genetic variation and developmental stability in quantitative traits. Proc. Natl. Acad. Sci. USA 2008, 105, 2963–2968. [Google Scholar]
- Song, H.M.; Wang, H.Z.; Xu, X.B. Overexpression of AtHsp90.3 in Arabidopsis thaliana impairs plant tolerance to heavy metal stress. Biol. Plant 2012, 56, 197–199. [Google Scholar]
- Lohmann, C.; Eggers-Schumacher, G.; Wunderlich, M.; Schöffl, F. Two different heat shock transcription factors regulate immediate early expression of stress genes in Arabidopsis. Mol. Genet. Genomics 2004, 271, 11–21. [Google Scholar]
- Xu, Z.S.; Chen, M.; Li, L.C.; Ma, Y.Z. Functions and application of the AP2/ERF transcription factor family in crop improvement. J. Integr. Plant Biol 2011, 53, 570–585. [Google Scholar]
- Shirasu, K.; Schulze-Lefert, P. Complex formation, promiscuity and multi-functionality: Protein interactions in disease-resistance pathways. Trends Plant Sci 2003, 8, 252–258. [Google Scholar]
- Takahashi, A.; Casais, C.; Ichimura, K.; Shirasu, K. HSP90 interacts with RAR1 and SGT1, and is essential for RPS2-mediated disease resistance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2003, 100, 11777–11782. [Google Scholar]
- Hubert, D.A.; Tornero, P.; Belkhadir, Y.; Krishna, P.; Takahashi, A.; Shirasu, K.; Dangl, J.L. Cytosolic HSP90 associates with and modulates the Arabidopsis RPM1 plant disease resistance protein. EMBO J 2003, 22, 5679–5689. [Google Scholar]
- Lu, R.; Malcuit, I.; Moffett, P.; Ruiz, M.T.; Peart, J.; Wu, A.J.; Rathjen, J.P.; Bendahmane, A.; Day, L.; Baulcombe, D.C. High throughput virus-induced gene silencing implicates heat shock protein 90 in plant disease resistance. EMBO J 2003, 22, 5690–5699. [Google Scholar]
- Liu, Y.; Burch-Smith, T.; Schiff, M.; Feng, S.; Dinesh-Kumar, S.P. Molecular chaperone Hsp90 associates with resistance protein N and its signaling proteins SGT1 and Rar1 to modulate an innate immune response in plants. J. Biol. Chem 2004, 279, 2101–2108. [Google Scholar]
- Hein, I.; Barciszewska-Pacak, M.; Hrubikova, K.; Williamson, S.; Dinesen, M.; Soenderby, I.E.; Sundar, S.; Jarmolowski, A.; Shirasu, K.; Lacomme, C. Virus-induced gene silencing-based functional characterization of genes associated with powdery mildew resistance in barley. Plant Physiol 2005, 138, 2155–2164. [Google Scholar]
- Scofield, S.R.; Huang, L.; Brandt, A.S.; Gill, B.S. Development of a virus-induced gene-silencing system for hexaploid wheat and its use in functional analysis of the Lr21-mediated leaf rust resistance pathway. Plant Physiol 2005, 138, 2165–2173. [Google Scholar]
- Wang, G.F.; Wei, X.; Fan, R; Zhou, H; Wang, X.; Yu, C.; Dong, L.; Dong, Z.; Wang, X.; Kang, Z.; et al. Molecular analysis of common wheat genes encoding three types of cytosolic heat shock protein 90 (Hsp90): Functional involvement of cytosolic Hsp90s in the control of wheat seedling growth and disease resistance. New Phytol. 2011, 191, 418–431. [Google Scholar]
- Bhattarai, K.K.; Li, Q.; Liu, Y.; Dinesh-Kumar, S.P.; Kaloshioan, I. The Mi-1-mediated pest resistance requires Hsp90 and Sgt1. Plant Physiol 2007, 144, 312–323. [Google Scholar]
- Liu, D.L.; Zhang, X.X.; Cheng, Y.X.; Takano, T.; Liu, S.K. Cloning and characterization of the rHsp90 gene in rice (Oryza sativa. L) under environmental stress. Mol. Plant Breed 2006, 4, 317–322. [Google Scholar]
- Yamada, K.; Fukao, Y.; Hayashi, M.; Fukazawa, M.; Suzuki, I.; Nishimura, M. Cytosolic HSP90 regulates the heat shock response that is responsible for heat acclimation in Arabidopsis thaliana. J. Biol. Chem 2007, 282, 37794–37804. [Google Scholar]
- Nishizawa-Yokoi, A.; Tainaka, H.; Yoshida, E.; Tamoi, M.; Yabuta, Y.; Shigeoka, S. The 26S proteasome function and Hsp90 activity involved in the regulation of HsfA2 expression in response to oxidative stress. Plant Cell Physiol 2010, 51, 486–496. [Google Scholar]
- McLellan, C.A.; Turbyville, T.J.; Wijeratne, E.M.; Kerschen, A.; Vierling, E.; Queitsch, C.; Whitesell, L.; Gunatilaka, A.A. A rhizosphere fungus enhances Arabidopsis thermotolerance through production of an HSP90 inhibitor. Plant Physiol 2007, 145, 174–182. [Google Scholar]
- Tominaga, H.; Coury, D.A.; Amano, H.; Miki, W.; Kakinuma, M. cDNA cloning and expression analysis of two heat shock protein genes, Hsp90 and Hsp60, from a sterile Ulva pertusa (Ulvales, Chlorophyta). Fish Sci 2012, 78, 415–429. [Google Scholar]
- Liu, D.; Zhang, X.; Cheng, Y.; Takano, T.; Liu, S. rHsp90 gene expression in response to several environmental stresses in rice (Oryza sativa L.). Plant Physiol. Bioch 2006, 44, 380–386. [Google Scholar]
- Hawle, P.; Horst, D.; Bebelman, J.P.; Yang, X.X.; Siderius, M.; van der Vies, S.M. Cdc37p is required for stress-induced high-osmolarity glycerol and protein kinase C mitogen-activated protein kinase pathway functionality by interaction with Hog1p and Slt2p (Mpk1p). Eukaryot. Cell 2007, 6, 521–532. [Google Scholar]
- Takabatake, R.; Ando, Y.; Seo, S.; Katou, S.; Tsuda, S.; Ohashi, Y.; Mitsuhara, I. MAP kinases function downstream of HSP90 and upstream of mitochondria in TMV resistance gene N-mediated hypersensitive cell death. Plant Cell Physiol 2007, 48, 498–510. [Google Scholar]
- Meiri, D.; Breiman, A. Arabidopsis ROF1 (FKBP62) modulates thermotolerance by interacting with HSP90.1 and affecting the accumulation of HsfA2-regulated sHSPs. Plant J 2009, 59, 387–399. [Google Scholar]
- Meiri, D.; Tazat, K.; Cohen-Peer, R.; Farchi-Pisanty, O.; Aviezer-Hagai, K.; Avni, A.; Breiman, A. Involvement of Arabidopsis ROF2 (FKBP65) in thermotolerance. Plant Mol. Biol 2010, 72, 191–203. [Google Scholar]
- Song, H.; Zhao, R.; Fan, P.; Wang, X.; Chen, X.; Li, Y. Overexpression of AtHsp90.2, AtHsp90.5 and AtHsp90.7 in Arabidopsis thaliana enhances plant sensitivity to salt and drought stresses. Planta 2009, 229, 955–964. [Google Scholar]

| Chaperones and relatives: | Transcription factors: |
|---|---|
|
|
| Kinases: | Others: |
|
|
| Nomenclature | Amino acids | Chromosome | Intracellular localization |
|---|---|---|---|
| AtHSP90.5 | 780 | II | Chloroplast stroma |
| AtHSP90.6 | 799 | III | Mitochondrial matrix space |
| AtHSP90.7 | 823 | IV | Endoplasmic reticulum |
| AtHSP90.1 | 705 | V | Cytoplasm |
| AtHSP90.4 | 699 | V | Nucleus |
| AtHSP90.3 | 668 | V | Nucleus |
| AtHSP90.2 | 728 | V | Nucleus |
| Nomenclature | Amino acids | Chromosome | Intracellular localization |
|---|---|---|---|
| Glyma08g44590.1 | 699 | VIII | Nucleus and plasma membrane |
| Glyma18g08220.1 | 702 | XVIII | Nucleus and plasma membrane |
| Glyma14g01530.1 | 700 | XIV | Nucleus and plasma membrane |
| Glyma02g47210.1 | 702 | II | Nucleus and plasma membrane |
| Glyma09g24410.1 | 699 | IX | Nucleus and plasma membrane |
| Glyma16g29750.1 | 699 | XVI | Nucleus and plasma membrane |
| Glyma08g03690.1 | 712 | VIII | Nucleus and plasma membrane |
| Glyma14g40320.1 | 847 | XIV | Endoplasmic reticulum |
| Glyma14g40320.2 | 816 | XIV | Endoplasmic reticulum |
| Glyma14g40320.4 | 727 | XIV | Nucleus and endoplasmic reticulum (lumen) |
| Glyma17g37820.1 | 814 | XVII | Endoplasmic reticulum |
| Glyma02g13790.1 | 794 | II | Chloroplast stroma and mitochondrial matrix space |
| Glyma02g47580.1 | 791 | II | Mitochondrial matrix space and nucleus |
| Glyma01g09310.1 | 793 | I | chloroplast stroma and mitochondrial matrix space |
| Glyma14g01100.1 | 797 | XIV | Mitochondrial matrix space and nucleus |
| Nomenclature | Amino acids | Chromosome | Intracellular localization |
|---|---|---|---|
| Os04g01740.1 | 703 | IV | Nucleus |
| Os06g50300.1 | 812 | VI | Endoplasmic reticulum |
| Os08g38086.3 | 761 | VIII | Mitochondrial matrix space |
| Os08g39140.1 | 699 | VIII | Nucleus |
| Os09g29840.1 | 791 | IX | Mitochondrial matrix space |
| Os09g30412.1 | 699 | IX | Nucleus |
| Os09g30418.1 | 830 | IX | Nucleus |
| Os09g36420.1 | 1046 | IX | Nucleus |
| Os12g32986.1 | 811 | XII | Vacuole |
| Nomenclature | Amino acids | Chromosome | Intracellular localization |
|---|---|---|---|
| Zm2G012631_T01 | 699 | II | Nucleus and mitochondrial matrix space |
| Zm2G112165_T01 | 698 | II | Nucleus and mitochondrial matrix space |
| Zm2G069651_T01 | 699 | II | Nucleus and mitochondrial matrix space |
| Zm5G833699_T01 | 714 | V | Nucleus and mitochondrial matrix space |
| Zm2G141931_T01 | 804 | II | Endoplasmic reticulum and vacuole |
| Zm2G141931_T03 | 710 | II | Endoplasmic reticulum and vacuole |
| Zm2G399073_T02 | 1001 | II | Endoplasmic reticulum |
| Zm2G399073_T03 | 808 | II | Endoplasmic reticulum |
| Zm2G141931_T04 | 667 | II | Endoplasmic reticulum |
| Zm2G002220_T01 | 793 | II | Chloroplast stroma and chloroplast thylakoid membrane |
| Zm5G813217_T01 | 758 | V | Nucleus and chloroplast stroma |
| Zm5G813217_T03 | 708 | V | Nucleus and mitochondrial matrix space |
© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Xu, Z.-S.; Li, Z.-Y.; Chen, Y.; Chen, M.; Li, L.-C.; Ma, Y.-Z. Heat Shock Protein 90 in Plants: Molecular Mechanisms and Roles in Stress Responses. Int. J. Mol. Sci. 2012, 13, 15706-15723. https://doi.org/10.3390/ijms131215706
Xu Z-S, Li Z-Y, Chen Y, Chen M, Li L-C, Ma Y-Z. Heat Shock Protein 90 in Plants: Molecular Mechanisms and Roles in Stress Responses. International Journal of Molecular Sciences. 2012; 13(12):15706-15723. https://doi.org/10.3390/ijms131215706
Chicago/Turabian StyleXu, Zhao-Shi, Zhi-Yong Li, Yang Chen, Ming Chen, Lian-Cheng Li, and You-Zhi Ma. 2012. "Heat Shock Protein 90 in Plants: Molecular Mechanisms and Roles in Stress Responses" International Journal of Molecular Sciences 13, no. 12: 15706-15723. https://doi.org/10.3390/ijms131215706
APA StyleXu, Z.-S., Li, Z.-Y., Chen, Y., Chen, M., Li, L.-C., & Ma, Y.-Z. (2012). Heat Shock Protein 90 in Plants: Molecular Mechanisms and Roles in Stress Responses. International Journal of Molecular Sciences, 13(12), 15706-15723. https://doi.org/10.3390/ijms131215706




