Pyrosequencing-Based Transcriptome Analysis of the Asian Rice Gall Midge Reveals Differential Response during Compatible and Incompatible Interaction
Abstract
:1. Introduction
2. Results and Discussion
2.1. Pyrosequencing and Assembly
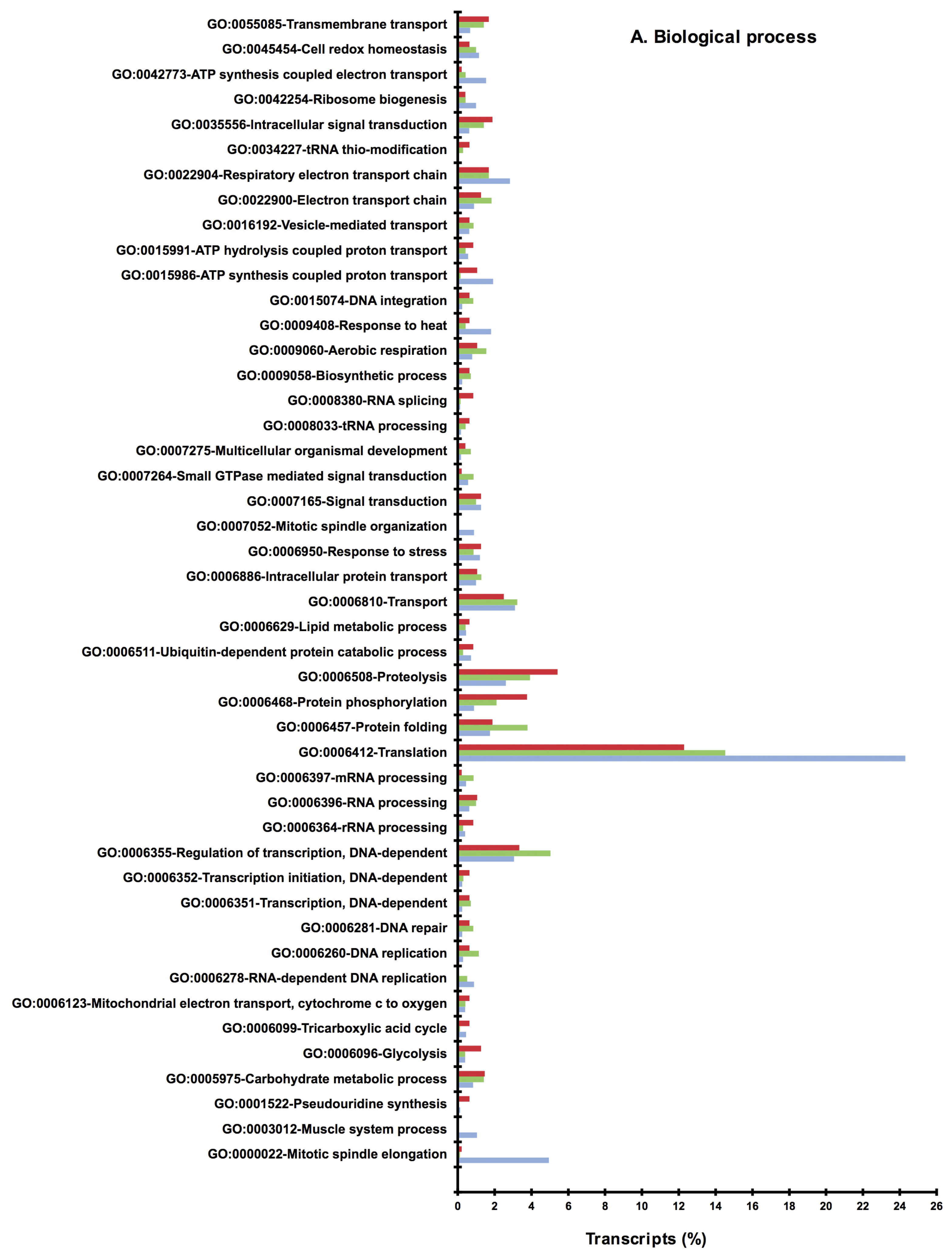
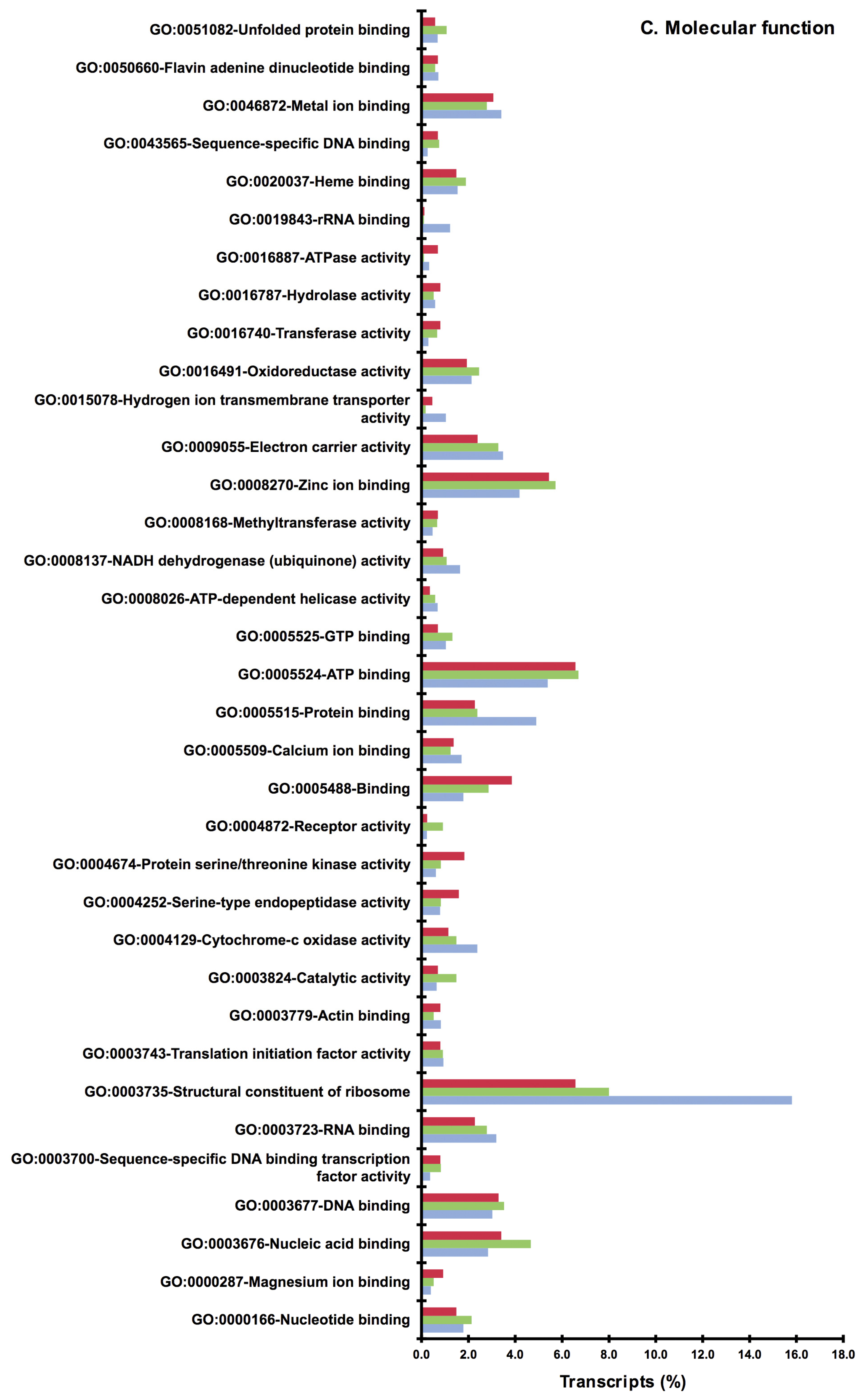
2.2. BLAST and Gene Ontology Analysis
2.3. Metabolic Pathway Analysis
2.4. Genes of Interest
2.4.1. Immune Regulatory Proteins
2.4.2. Proteases
2.4.3. Protein Kinases
2.4.4. Genes Involved in Apoptosis
2.4.5. Genes Related to Reactive Oxygen Species (ROS) Pathways
2.5. Detection of Molecular Markers
2.6. Comparative Analyses of the Asian Rice Gall Midge Transcripts with the Hessian Fly
2.7. Comparative Analyses of Transcripts
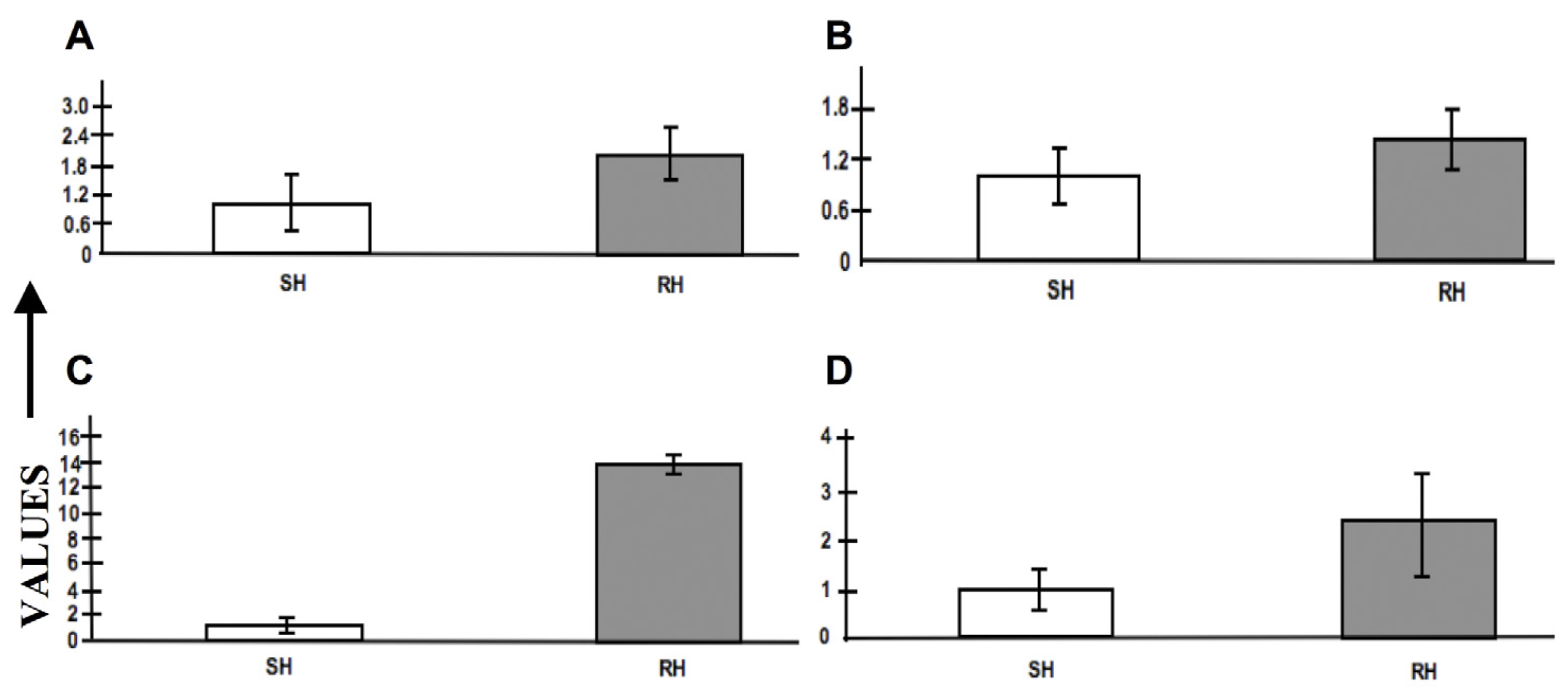
2.8. Expression Profiling by RT-PCR
3. Experimental Section
3.1. Collection of Orseolia oryzae Larvae and RNA Isolation
3.2. RNA Isolation and 454 Sequencing
3.3. Transcript Assembly and Data Processing
3.4. Real-time PCR and Statistical Analyses
4. Conclusions
Supplementary Materials
ijms-13-13079-s001.pdfAcknowledgments
- Conflict of InterestThe authors declare no conflict of interest.
References
- Despres, L.; David, J.P.; Gallet, C. The evolutionary ecology of insect resistance to plant chemicals. Trends Ecol. Evol 2007, 22, 298–308. [Google Scholar]
- Raman, A.; Burckhardt, D.; Harris, K.M. Biology and adaptive radiation in the gall-inducing Cecidomyiidae (Insecta Diptera) and Calophyidae (Insecta Hemiptera) on Mangifera indica (Anacardiaceae) in the Indian subcontinent. Trop. Zool 2009, 22, 27–56. [Google Scholar]
- Harris, M.O.; Stuart, J.J.; Mohan, M.; Nair, S.; Lamb, R.J.; Rohfritsch, O. Grasses and gall midges: Plant defense and insect adaptation. Annu. Rev. Entomol 2003, 48, 549–577. [Google Scholar]
- Bentur, J.S.; Pasalu, I.C.; Sarma, N.P.; Prasad Rao, U.; Mishra,, B. Gall Midge Resistance in Rice; Directorate of Rice Research: Hyderabad, India, 2003; p. 22. [Google Scholar]
- Bentur, J.S.; Srinivasan, T.E.; Kalode, M.B. Occurrence of a virulent rice gall midge (GM) Orseolia oryzae Wood-Mason biotype (?) in Andhra Pradesh, India. Int. Rice Res. Newslett 1987, 12, 33–34. [Google Scholar]
- Sardesai, N.; Rajyashri, K.R.; Behura, S.K.; Nair, S.; Mohan, M. Genetic, physiological and molecular interactions of rice and its major dipteran pest, gall midge. Plant Cell Tissue Organ Cult 2001, 64, 115–131. [Google Scholar]
- Bentur, J.S.; Kalode, M.B. Hypersensitive reaction and induced resistance in rice against the Asian rice gall midge (Orseolia oryzae). Entomol. Exp. Appl 1996, 78, 77–81. [Google Scholar]
- Himabindu, K.; Suneetha, K.; Sama, V.S.A.K.; Bentur, J.S. A new rice gall midge resistance gene in the breeding line CR57-MR1523, mapping with flanking markers and development of NILs. Euphytica 2010, 174, 179–187. [Google Scholar]
- Sinha, D.K.; Bentur, J.S.; Nair, S. Compatible interaction with its rice host leads to enhanced expression of gamma subunit of oligosaccharyl transferase (OoOST) in the Asian rice gall midge (Orseolia oryzae). Insect Mol. Biol 2011, 20, 567–575. [Google Scholar]
- Sinha, D.K.; Lakshmi, M.; Anuradha, G.; Rahman, S.J.; Siddiq, E.A.; Bentur, J.S.; Nair, S. Serine proteases-like genes in the rice gall midge show differential expression in compatible and incompatible interactions with rice. Int. J. Mol. Sci 2011, 12, 2842–2852. [Google Scholar]
- Pauchet, Y.; Wilkinson, P.; Vogel, H.; Nelson, D.R.; Reynolds, S.E.; Heckel, D.G.; Ffrench-Constant, R.H. Pyrosequencing the Manduca sexta larval midgut transcriptome: Messages for digestion, detoxification and defence. Insect Mol. Biol 2010, 19, 61–75. [Google Scholar]
- Leshkowitz, D.; Gazit, S.; Reuveni, E.; Ghanim, M.; Czosnek, H.; McKenzie, C.; Shatters, R.L., Jr; Brown, J.K. Whitefly (Bemisia tabaci) genome project: Analysis of sequenced clones from egg, instar, and adult (viruliferous and non-viruliferous) cDNA libraries. BMC Genomics 2006, 7, 79. [Google Scholar]
- Bai, X.; Zhang, W.; Orantes, L.; Jun, T.-H.; Mittapalli, O.; Mian, M.A.; Michel, A.P. Combining next-generation sequencing strategies for rapid molecular resource development from an invasive aphid species, Aphis glycines. PLoS One 2010, 5, e11370. [Google Scholar]
- Mittapalli, O.; Bai, X.; Mamidala, P.; Rajarapu, S.P.; Bonello, P.; Herms, D.A. Tissue-specific transcriptomics of the exotic invasive insect pest emerald ash borer (Agrilus planipennis). PLoS One 2010, 5, e13708. [Google Scholar]
- Margulies, M.; Egholm, M.; Altman, W.E.; Attiya, S.; Bader, J.S.; Bemben, L.A.; Berka, J.; Braverman, M.S.; Chen, Y.-J.; Chen, Z.; et al. Genome sequencing in open microfabricated high density picoliter reactors. Nature 2005, 437, 376–380. [Google Scholar]
- UniRef Databases. Available online: http://www.uniprot.org/uniref/ accessed on 28 January 2012.
- Suzek, B.E.; Huang, H.; McGarvey, P.; Mazumder, R.; Wu, C.H. UniRef: Comprehensive and non-redundant UniProt reference clusters. Bioinformatics 2007, 43, 1282–1288. [Google Scholar]
- KEGG Automatic Annotation Server (KAAS). Available online: http://www.genome.jp/tools/kaas/ accessed on 12 December 2011.
- Saltzmann, K.D.; Giovanini, M.P.; Zheng, C.; Williams, C.E. Virulent hessian fly larvae manipulate the free amino acid content of host wheat plants. J. Chem. Ecol 2008, 34, 1401–1410. [Google Scholar]
- Pace, K.E.; Baum, L.G. Insect galectin: Roles in immunity and development. Glycoconj. J 2004, 19, 604–617. [Google Scholar]
- Vasta, G.R. Galectins as pattern recognition receptors: Structure, function and evolution. Adv. Exp. Med. Biol 2012, 946, 21–36. [Google Scholar]
- Sideri, M.; Tsakas, S.; Markoutsa, E.; Lampropoulou, M; Marmaras, V.J. Innate immunity in insects: Surface-associated dopa decarboxylase-dependent pathways regulate phagocytosis, nodulation and melanization in medfly haemocytes. Immunology 2008, 123, 528–537. [Google Scholar]
- Broadway, R.M. Dietary regulation of serine proteinases that are resistant to serine proteinase inhibitors. J. Insect Physiol 1997, 43, 855–874. [Google Scholar]
- Zhang, S.; Shukle, R.; Mittapalli, O.; Zhu, Y.C.; Reese, J.C.; Wang, H.; Hua, B.Z.; Chen, M.S. The gut transcriptome of a gall midge, Mayetiola destructor. J. Insect Physiol 2010, 56, 1198–1206. [Google Scholar]
- Jongsma, M.A.; Bakker, P.L.; Peters, J.; Bosch, D.; Stiekema, W.J. Adaptation of Spodoptera exigua larvae to plant proteinase inhibitors by induction of gut proteinase activity insensitive to inhibition. Proc. Natl. Acad. Sci. USA 1995, 92, 8041–8045. [Google Scholar]
- Brioschi, D.; Nadalini, L.D.; Bengtson, M.H.; Sogayar, M.C.; Moura, D.S.; Silva-Filho, M.C. General upregulation of Spodoptera frugiperda trypsins and chymotrypsins allows its adaptation to soybean proteinase inhibitor. Insect Biochem. Mol. Biol 2007, 37, 1283–1290. [Google Scholar]
- Bonshtien, A.; Lev, A.; Gibly, A.; Debbie, P.; Avni, A.; Sessa, G. Molecular properties of the Xanthomonas AvrRxv effector and global transcriptional changes determined by its expression in resistant tomato plants. Mol. Plant Microbe Int 2005, 18, 300–310. [Google Scholar]
- Jia, Y.; McAdams, S.A.; Bryan, G.T.; Hershey, H.P.; Valent, B. Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J 2000, 19, 4004–4014. [Google Scholar]
- Welchman, D.P.; Aksoy, S.; Jiggins, F.; Lemaitre, B. Insect immunity: From pattern recognition to symbiont-mediated host defense. Cell Host Microbe 2009, 6, 107–114. [Google Scholar]
- Natori, S.; Shiraishi, H.; Hori, S.; Kobayashi, A. The roles of Sarcophaga defense molecules in immunity and metamorphosis. Dev. Comp. Immunol 1999, 23, 317–328. [Google Scholar]
- Matsumoto, I.; Emori, Y.; Abe, K.; Arai, S. Characterization of a gene family encoding cysteine proteinases of Sitophilus zeamais (Maize Weevil), and analysis of the protein distribution in various tissues including alimentary tract and germ cells. J. Biochem 1997, 121, 464–476. [Google Scholar]
- Yamamoto, Y.; Takimoto, K.; Izumi, S.; Toriyama-Sakurai, M; Kageyama, T.; Takahashi, S.Y. Molecular cloning and sequencing of cDNA that encodes cysteine proteinase in the eggs of the silkmoth, Bombyx mori. J. Biochem 1994, 116, 1330–1335. [Google Scholar]
- Fujii-Taira, I.; Tanaka, Y.; Homma, K.J.; Natori, S. Hydrolysis and synthesis of substrate proteins for cathepsin L in the brain basement membranes of Sarcophaga during metamorphosis. J. Biochem 2000, 128, 539–542. [Google Scholar]
- Zhu, Y.C.; Liu, X.; Maddur, A.A.; Oppert, B.; Chen, M.S. Cloning and characterization of chymotrypsin- and trypsin-like cDNAs from the gut of the Hessian fly [Mayetiola destructor (Say)]. Insect Biochem. Mol. Biol 2005, 35, 23–32. [Google Scholar]
- Sharifpoor, S.; Nguyen, A.N.; Young, J.Y.; Dyk, D.V.; Friesen, H.; Douglas, A.C.; Kurat, C.F; Chong, Y.T.; Founk, K.; Moses, A.M. A quantitative literature-curated gold standard for kinase-substrate pairs. Genome Biol 2011, 12, R39. [Google Scholar]
- Manning, G. Genomic overview of protein kinases. In WormBook: The Online Review of C. elegans Biology; WormBook Research: Pasadena, CA, USA, 2005. [Google Scholar]
- Hammond-Kosack, K.; Urban, M.; Baldwin, T.; Daudi, A.; Rudd, J.; Keon, J.; Lucas, J.; Maguire, K.; Kornyukhin, D.; Jing, H.-C.; et al. Plant Pathogens: How Can Molecular Genetic Information on Plant Pathogens Assist in Breeding Disease Resistant Crops. Proceedings of the 4th International Crop Science Congress, Brisbane, Australia, 26 September–1 October 2004.
- Belvin, M.P.; Anderson, K.V. A conserved signaling pathway: The Drosophila Toll-Dorsal pathway. Annu. Rev. Cell Dev. Biol 1996, 12, 393–416. [Google Scholar]
- Arai, K.; Lee, S.R.; van Leyen, K.; Kurose, H.; Lo, E.H. Involvement of ERK MAP kinase in endoplasmic reticulum stress in SH-SY5Y human neuroblastoma cells. J. Neurochem 2004, 89, 232–239. [Google Scholar]
- Rybczynski, R.; Bell, S.C.; Gilbert, L.I. Activation of an extracellular signal-regulated kinase (ERK) by the insect prothoracicotropic hormone. Mol. Cell Endocrinol 2001, 184, 1–11. [Google Scholar]
- Pfister, T.D.; Storey, K.B. Insect freeze tolerance: Roles of protein phosphatases and protein kinase A. Insect Biochem. Mol. Biol 2006, 36, 18–24. [Google Scholar]
- Mittapalli, O.; Shukle, R.H. Molecular characterization and responsive expression of a defender against apoptotic cell death homologue from the Hessian fly, Mayetiola destructor. Comp. Biochem. Physiol 2008, 149, 517–523. [Google Scholar]
- Mittapalli, O.; Neal, J.J.; Shukle, R.H. Antioxidant defense response in a galling insect. Proc. Natl. Acad. Sci. USA 2007, 104, 1889–1894. [Google Scholar]
- Chen, M.S.; Liu, X.; Yang, Z.; Zhao, H.; Shukle, R.H.; Stuart, J.J.; Hulbert, S. Unusual conservation among genes encoding small secreted salivary gland proteins from a gall midge. BMC Evol. Biol 2010, 10, 296. [Google Scholar]
- Zhang, F.; Hongyan, G.; Huajun, Z.; Tong, Z.; Yijun, Z.; Wang, S.; Fang, R.; Qian, W.; Chen, X. Massively parallel pyrosequencing-based transcriptome analyses of small brown planthopper (Laodelphax striatellus), a vector insect transmitting rice stripe virus (RSV). BMC Genomics 2010, 11, 303. [Google Scholar]
- Behura, S.K.; Sahu, S.C.; Mohan, M.; Nair, S. Wolbachia in the Asian rice gall midge, Orseolia oryzae (Wood-Mason): Correlation between host mitotypes and infection status. Insect Mol. Biol 2001, 10, 163–171. [Google Scholar]
- Lakshmi, P.V.; Amudhan, S.; Himabindu, K.; Cheralu, C.; Bentur, J.S. A new biotype of the Asian rice gall midge Orseolia oryzae (Diptera: Cecidomyiidae) characterized from the Warangal population in Andhra Pradesh, India. Int. J. Trop. Insect Sci 2006, 26, 207–211. [Google Scholar]
- MIRA 3. Available online: http://www.chevreux.org/projects_mira.html accessed on 30 April 2011.
- Blast2GO. Available online: http://www.blast2go.org accessed on 13 February 2012.
- Microsatellite identification tool (MISA). Available online: http://pgrc.ipk-gatersleben.de/misa/ accessed on 21 May 2012.
- Primer3. Available online: http://frodo.wi.mit.edu/ accessed on 8 July 2011.
- Vandesompele, J.; de Preter, K.; Pattyn, F.; Poppe, B.; van Roy, N.; de Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 2002, 3, research0034.1–research0034.11. [Google Scholar]
- Sokal, R.R.; Rohlf, F.J. Biometry: The Principles and Practice of Statistics in Biological Research, 3rd ed; W H Freeman and Co: New York, NY, USA, 1995; p. 887. [Google Scholar]
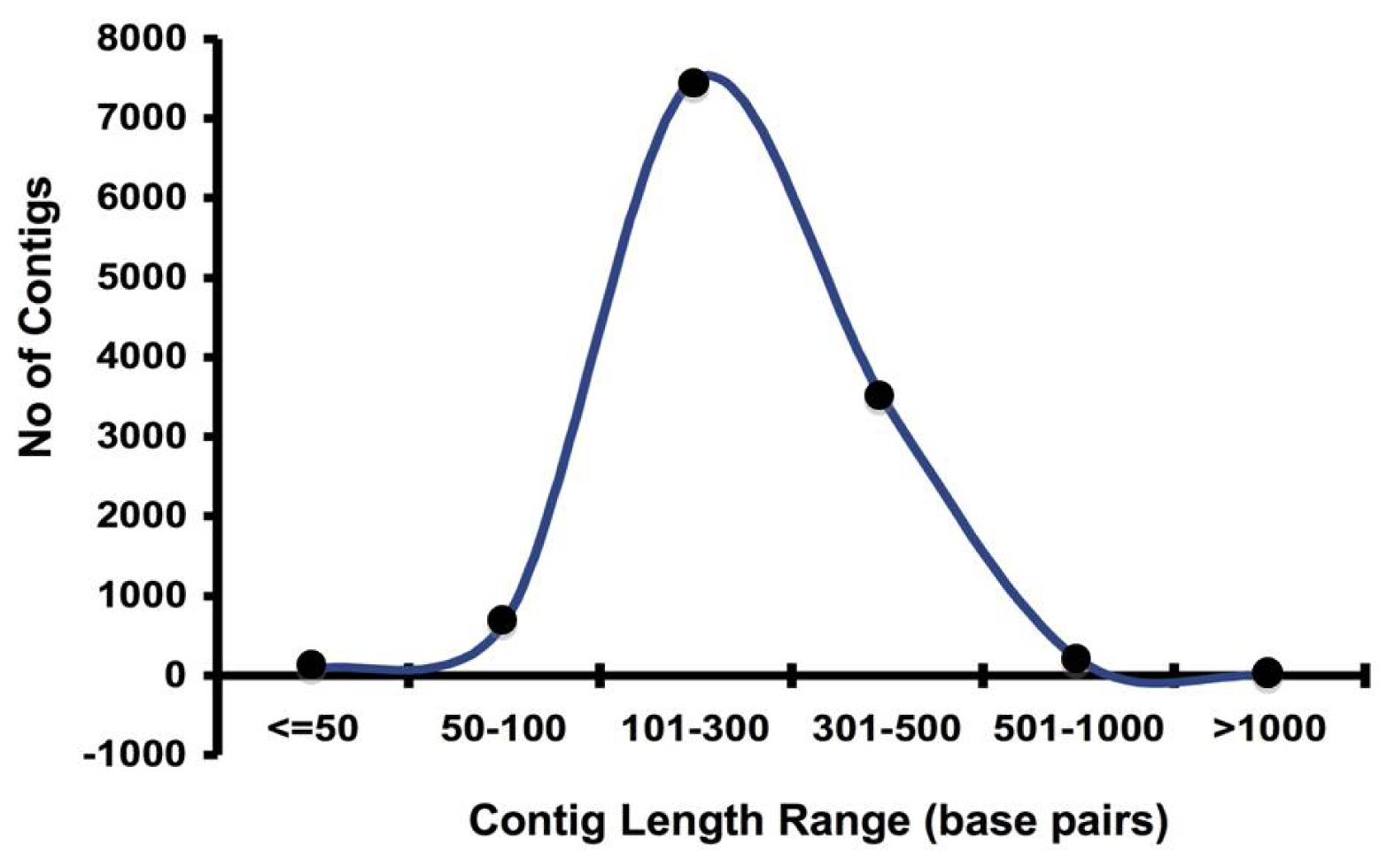
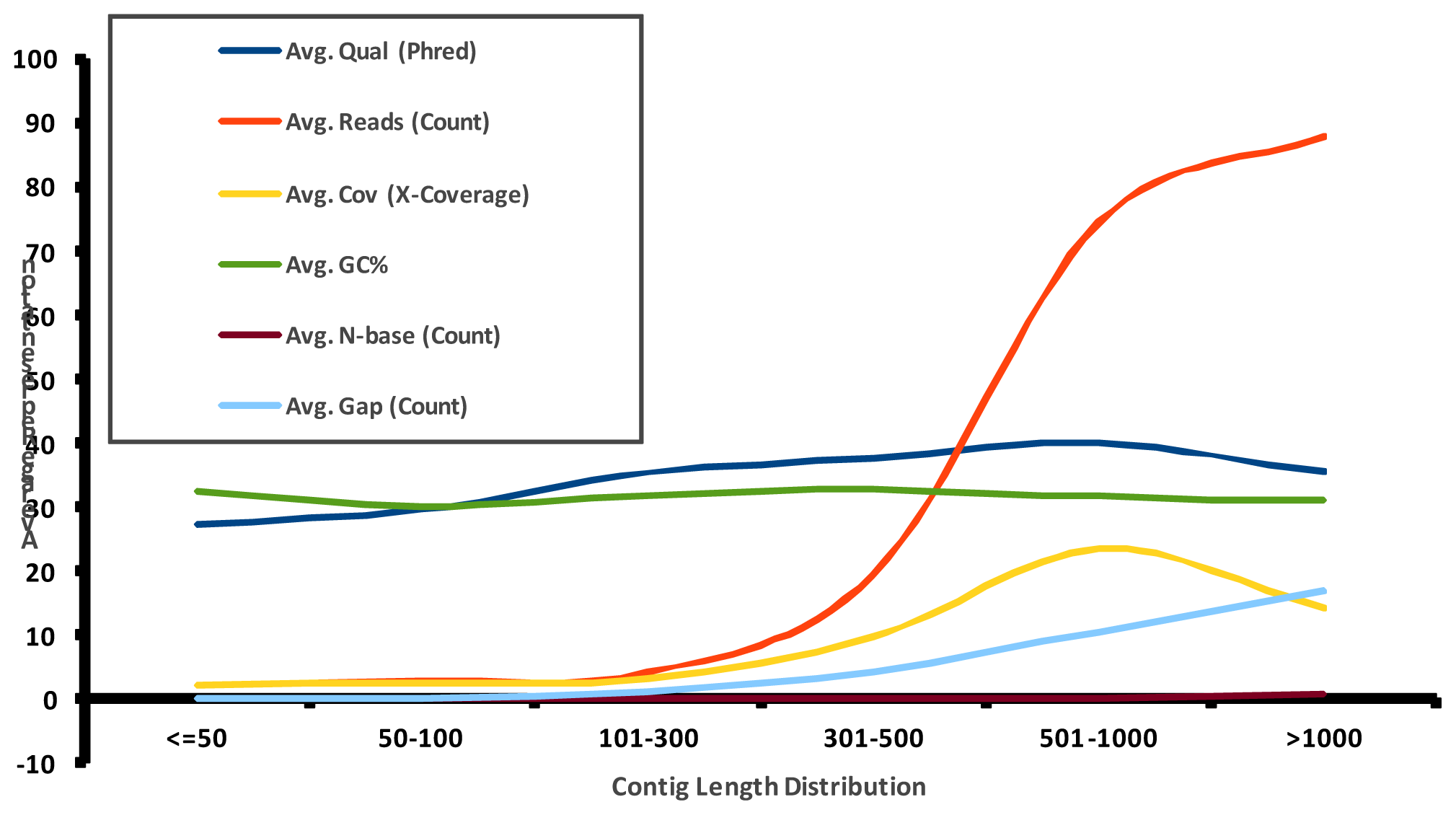
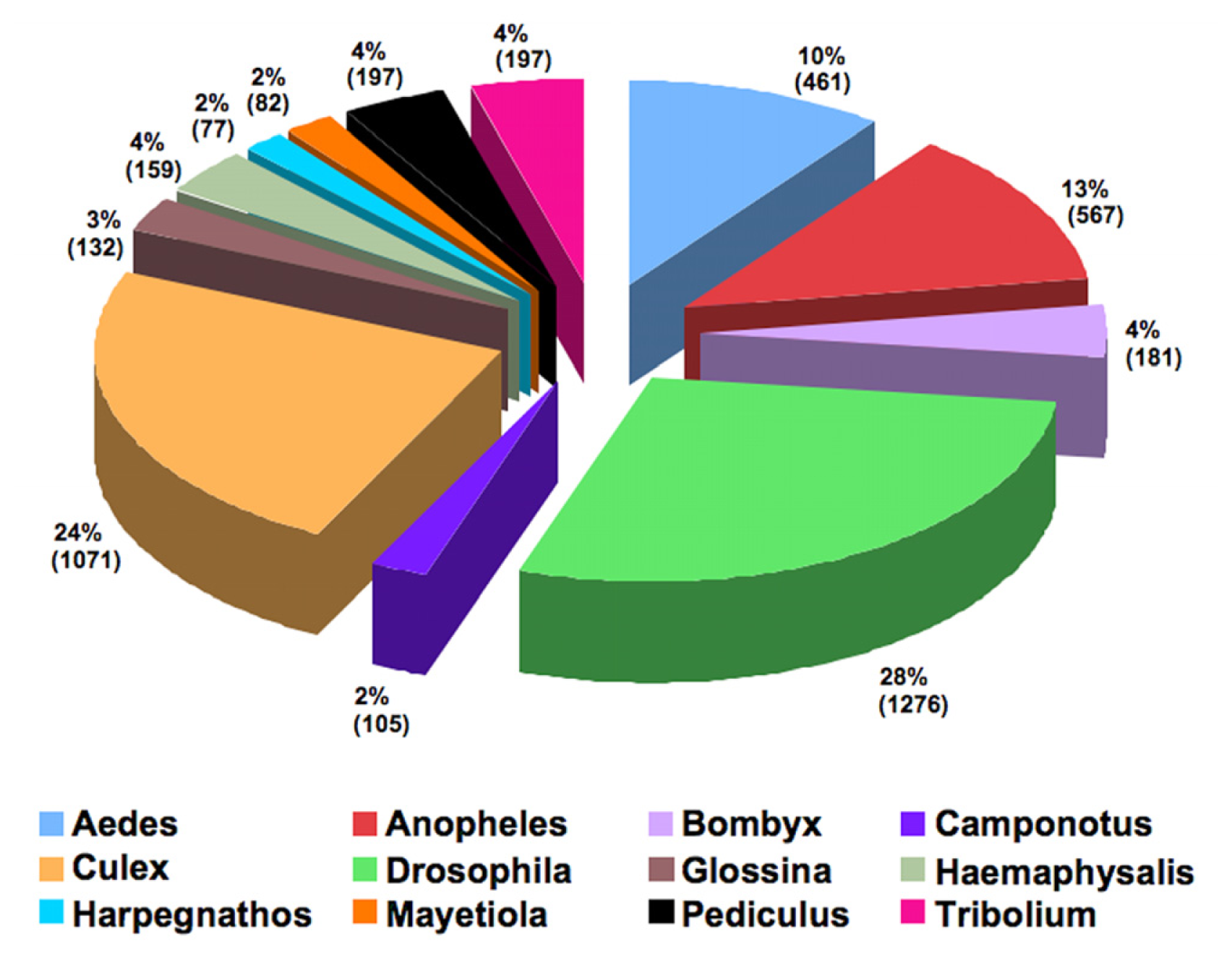
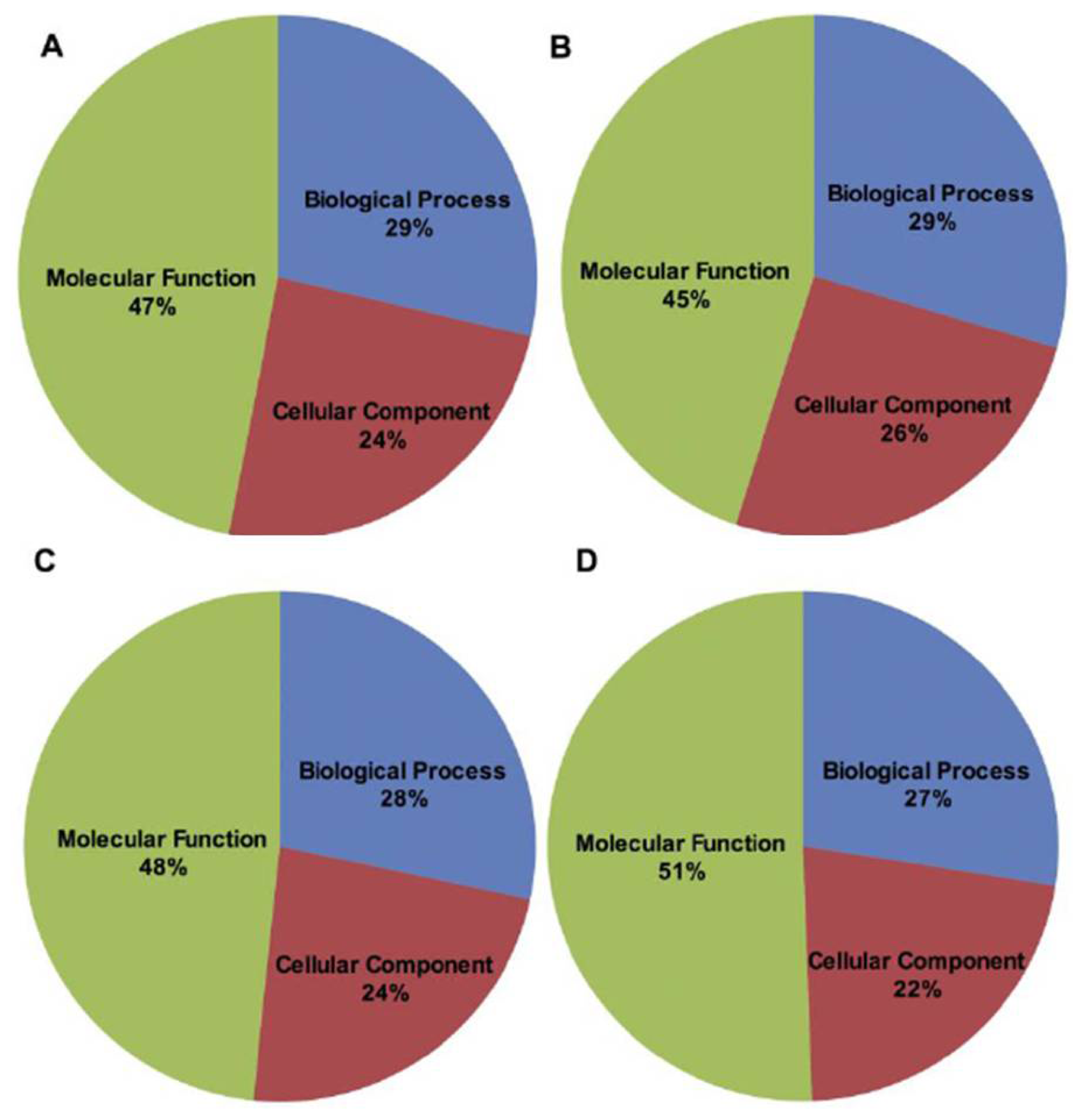


| Virulent GMB4 | Avirulent GMB4 | |
|---|---|---|
| Total number of sequence (bp) after filtering | 79,028 (76,963) * | 85,395 (84,118) * |
| High quality reads (%) ** | 70.25 | 72.44 |
| Average reads length (bp) | 242 | 236 |
| Sequence length (%) | - | - |
| <100 | 3.01 | 1.67 |
| 101–500 | 96.63 | 98.03 |
| 501–1000 | 0.35 | 0.30 |
| 1001–1500 | 0.01 | 0 |
| Number of contigs | 9,272 | 9,526 |
| Number of singletons | 21,764 | 24,625 |
| Functions | Code of ESTs | E-value | Similarity |
|---|---|---|---|
| Melanization | |||
| ICGEB_c3627 | 4 × 10−48 | Dihydropteridine reductase | |
| ICGEB_rep_c3461 | 50 | Prophenoloxidase | |
| FKB78SY06DOCUN | 7 × 10−18 | Serpin | |
| FKB78SY16JUF7R | 3 × 10−5 | Serpin 1 | |
| FKB78SY14ITCIU | 1.10 × 10−3 | Laccase-4 | |
| Immune regulatory proteins | |||
| FKB78SY09FO3VQ | 2 × 10−10 | Toll | |
| FKB78SY15I6036 | 2 × 10−9 | Intermediate in Toll signalling pathway | |
| ICGEB_rep_c1724 | 9 × 10−70 | Ubiquitin carrier protein | |
| FKB78SY10GAGBA | 1 × 10−14 | GH14989 | |
| ICGEB_rep_c1629 | 1 × 10−47 | Histone H2A | |
| ICGEB_rep_c599 | 2 × 10−8 | Transcription factor NFAt subunit NF45 | |
| ICGEB_rep_c25 | 6 × 10−22 | C-type lectin, galactose-binding | |
| ICGEB_c3769 | 1 × 10−5 | Galactose-specific C-type lectin | |
| ICGEB_c2381 | 2 × 10−32 | C-type lectin | |
| ICGEB_c10863 | 4 × 10−2 | Lysozyme c-1 | |
| FKB78SY15JHPUN | 6.2 × 10−3 | Argonaute 2 | |
| Proteases | |||
| ICGEB_rep_c638 | 6 × 10−25 | Cathepsin L precursor | |
| ICGEB_rep_c1661 | 6 × 10−9 | Putative gut cathepsin d-like aspartic protease | |
| ICGEB_c2413 | 8 × 10−9 | Cathepsin l-like cysteine proteinase | |
| ICGEB_rep_c250 | 3 × 10−6 | Cathepsin l-like cysteine proteinase CAL1 | |
| ICGEB_rep_c1725 | 1 × 10−39 | Chymotrypsin-like serine protease | |
| ICGEB_rep_c3018 | 4 × 10−39 | Serine protease P100 | |
| ICGEB_c3187 | 1 × 10−35 | Serine protease H2 | |
| ICGEB_c5978 | 1 × 10−22 | CLIP-domain serine protease subfamily D | |
| ICGEB_c2987 | 7 × 10−13 | Aspartic protease | |
| ICGEB_rep_c1876 | 3 × 10−11 | Digestive cysteine protease | |
| ICGEB_rep_c9287 | 6 × 10−11 | Signal peptide protease | |
| ICGEB_c4650 | 3 × 10−9 | Serine protease htra2 | |
| ICGEB_rep_c1661 | 6 × 10−9 | Putative gut cathepsin d-like aspartic protease | |
| ICGEB_c6152 | 2 × 10−8 | Lysosomal aspartic protease | |
| FKB78SY06DMP3X | 4 × 10−3 | Serine protease snake, putative | |
| Kinase | |||
| FKB78SY14IS5EN | 9 × 10−34 | Serine-threonine kinase receptor-associated protein | |
| FKB78SY14IKKDH | 5 × 10−30 | Dual specificity MAPKK4 | |
| ICGEB_c2935 | 2 × 10−26 | MAPK kinase 1-interacting protein 1 | |
| ICGEB_c8827 | 2 × 10−22 | Inositol polyphosphate multikinase | |
| ICGEB_c8292 | 6 × 10−22 | Casein kinase II subunit alpha | |
| FKB78SY09FSJ0Q | 9 × 10−16 | Serine-threonine kinase | |
| FKB78SY11GZ55Y | 3 × 10−18 | Src tyrosine kinase, putative | |
| ICGEB_c8214 | 6 × 10−20 | Nucleoside diphosphate kinase | |
| Apoptosis and cell death | |||
| ICGEB_c2475 | 7 × 10−13 | Caspase long class, Dronc-like | |
| FKB78SY07D8PWI | 4 × 10−14 | Inhibitor of apoptosis 2 protein | |
| ICGEB_c3186 | 1 × 10−28 | Defender against apoptotic cell death | |
| ICGEB_rep_c1321 | 3 × 10−6 | Programmed cell death protein 7 | |
| ICGEB_c10998 | 6 × 10−4 | Programmed cell death 4a | |
| ROS related genes | |||
| ICGEB_c8627 | 7 × 10−12 | Catalase | |
| ICGEB_rep_c11026 | 1 × 10−21 | Superoxide dismutase [Cu-Zn] | |
| ICGEB_rep_c991 | 6 × 10−40 | Glutathione S-transferase | |
| ICGEB_c5428 | 4 × 10−38 | Glutathione peroxidase | |
| ICGEB_c4435 | 6 × 10−11 | Cytochrome P450 | |
| Microsatellite Repeats | Number of loci |
|---|---|
| Dincucleotide | 2,380 |
| Trinucleotide | 1,205 |
| Tetranucleotide | 134 |
| Pentanucleotide | 56 |
| Hexanucleotide | 300 |
| Complex | 143 |
| Total | 4,218 |
| SNP type | Counts |
|---|---|
| Transition | |
| A-G | 282 |
| C-T | 120 |
| Transversion | |
| A-C | 300 |
| A-T | 390 |
| C-G | 49 |
| G-T | 101 |
| Total | 1242 |
| Others * | 1514 |
| Grand Total | 2756 |
| Specifically present in virulent GMB4 | Specifically present in avirulent GMB4 | Commonly present in both interaction | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Contig id | Similarity | CL | N | Contig id | Similarity | CL | N | Contig id | Similarity | CL | N | |
| Virulent | Avirulent | |||||||||||
| ICGEB_rep_c1075 | Zinc finger protein | 128 | 18 | ICGEB_c2372 | Serine-threonine protein phosphatase | 176 | 11 | ICGEB_rep_c6546 | Tropomyosin-2 | 164 | 366 | 449 |
| ICGEB_rep_c1403 | AGAP008060-PA | 302 | 10 | ICGEB_c3178 | Piopio protein | 281 | 8 | ICGEB_rep_c43 | Predicted protein | 464 | 133 | 484 |
| ICGEB_rep_c1433 | Putative uncharacterized protein | 187 | 9 | ICGEB_c2916 | Putative uncharacterized protein | 326 | 8 | ICGEB_rep_c128 | Ribosomal protein L37 | 251 | 470 | 375 |
| ICGEB_rep_c1962 | GF23525 | 80 | 8 | ICGEB_rep_c1359 | Pseudouridine synthase | 113 | 8 | ICGEB_rep_c21 | 60S ribosomal protein L24 | 232 | 319 | 277 |
| ICGEB_c2217 | Proteasome subunit beta type | 100 | 8 | ICGEB_c2037 | Putative uncharacterized protein | 287 | 7 | ICGEB_rep_c12 | ATP synthase subunit a | 429 | 211 | 358 |
| ICGEB_c2817 | Putative uncharacterized protein | 182 | 7 | ICGEB_rep_c4187 | Putative uncharacterized protein | 107 | 7 | ICGEB_rep_c2 | Midline fasciclin | 167 | 306 | 229 |
| ICGEB_c2608 | Ribosomal protein S25 | 96 | 7 | ICGEB_rep_c3796 | Ribosomal protein, L48, putative | 200 | 7 | ICGEB_rep_c24 | GH15515 | 257 | 240 | 186 |
| ICGEB_c2406 | FKBP-type peptidylprolyl cis-trans isomerase | 164 | 7 | ICGEB_c2559 | GF20993 | 173 | 6 | ICGEB_rep_c19 | 60S ribosomal protein L23 | 290 | 203 | 201 |
| ICGEB_c2935 | Mitogen-activated protein kinase kinase 1-interacting protein 1 | 284 | 7 | ICGEB_c2421 | GL24166 | 185 | 6 | ICGEB_rep_c10 | Cytochrome C oxidase subunit 1 | 229 | 238 | 194 |
| ICGEB_c2896 | Multiple coagulation factor deficiency protein 2-like protein | 222 | 7 | ICGEB_c2757 | AGAP004322-PA (Fragment) | 164 | 6 | ICGEB_rep_c82 | Cytochrome b | 224 | 217 | 223 |
| ICGEB_rep_c2089 | APAF1-interacting protein-like protein | 170 | 7 | ICGEB_rep_c4363 | GI18785 | 208 | 5 | ICGEB_rep_c10597 | 60S ribosomal protein L12 | 217 | 245 | 97 |
| ICGEB_rep_c2623 | Putative uncharacterized protein | 135 | 7 | ICGEB_c4321 | GJ10149 | 80 | 5 | ICGEB_rep_c54 | Ribosomal protein L19 | 326 | 212 | 140 |
| ICGEB_c2915 | Putative uncharacterized protein | 100 | 7 | ICGEB_c10901 | Putative uncharacterized protein | 95 | 5 | ICGEB_rep_c3468 | NAD-Hubiquinone oxidoreductase chain 4 | 347 | 193 | 155 |
| ICGEB_c2381 | C-type lectin | 212 | 7 | ICGEB_c2603 | Putative uncharacterized protein | 110 | 5 | ICGEB_rep_c25 | AGAP010193-PA | 1139 | 174 | 156 |
| ICGEB_c2477 | GA11576 | 206 | 7 | ICGEB_rep_c2725 | GA19828 | 269 | 5 | ICGEB_rep_c107 | GL12416 | 137 | 214 | 110 |
| S. No | Gene name | Primer name | Primer sequence (5′-3′) | Tm (°C) | Amplicon size (bp) |
|---|---|---|---|---|---|
| 1 | Cytochrome oxidase I | RTCytoc F | TGTAGGAATAGAAGTTGATACACGAGCTT | 60 | 185 |
| RTCytoc R | CTCCTGTCACTCCTCCAATAGTAAATAA | ||||
| 2 | Serine-threonine phosphatase | RTSTP F | TAAAGACATGCGAGGGTGAGAGT | 60 | 120 |
| RTSTP R | CAGCGACAGAAAATGGTGACA | ||||
| 3 | APAF1-interacting protein | RTAIP1 F | GTTCGCCGACACGGACTTTA | 60 | 104 |
| RTAIP1 R | TTCTTCATTTCCACAGCAATTCC | ||||
| 4 | Cyclophilin | RTCyclop F | GGTATTTTTGGATATGTCGTCGAA | 60 | 102 |
| RTCyclop R | GCTGCTGATTATCACATTACTTTGTG | ||||
| 5 | Tetraspanin 139 | RTTet139 F | TCACCATCCGAATGGATTCC | 60 | 129 |
| RTTet139 R | CCCGCTGCCAATCAATTCT | ||||
| 6 | Adenylate cyclase | RTAdcycl F | GAGGCCCGGCAAAGAAGA | 60 | 100 |
| RTAdcycl R | AGCGAGTGCAAATTCCACAAC | ||||
| 7 | MAPK interacting serine-threonine kinase | RTMAPKistk F | CTGAAAGCGAAAATGCCGATA | 60 | 100 |
| RTMAPKistk R | CTCAATTCACGTGCCGATTG | ||||
| 8 | C-type lectin | RTCTL F | CGGTGCCCACGAAAACTG | 60 | 131 |
| RTCTL R | GCACATATTTCAGAAGTGCATCATT | ||||
| 9 | Inositol polyphosphate multikinase | RTIPMK F | GAGAATGGGCCTATGTCAAAATG | 60 | 101 |
| RTIPMK R | CACAAGATTTTCGATGCCAAATAA | ||||
| 10 | Caspase | RTCasp F | AAACGAGTAGTGAAGGTGCAAACATA | 60 | 150 |
| RTCasp R | CGTGCGCATGTTCAGCTAAT | ||||
© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Sinha, D.K.; Nagaraju, J.; Tomar, A.; Bentur, J.S.; Nair, S. Pyrosequencing-Based Transcriptome Analysis of the Asian Rice Gall Midge Reveals Differential Response during Compatible and Incompatible Interaction. Int. J. Mol. Sci. 2012, 13, 13079-13103. https://doi.org/10.3390/ijms131013079
Sinha DK, Nagaraju J, Tomar A, Bentur JS, Nair S. Pyrosequencing-Based Transcriptome Analysis of the Asian Rice Gall Midge Reveals Differential Response during Compatible and Incompatible Interaction. International Journal of Molecular Sciences. 2012; 13(10):13079-13103. https://doi.org/10.3390/ijms131013079
Chicago/Turabian StyleSinha, Deepak Kumar, Javaregowda Nagaraju, Archana Tomar, Jagadish S. Bentur, and Suresh Nair. 2012. "Pyrosequencing-Based Transcriptome Analysis of the Asian Rice Gall Midge Reveals Differential Response during Compatible and Incompatible Interaction" International Journal of Molecular Sciences 13, no. 10: 13079-13103. https://doi.org/10.3390/ijms131013079
APA StyleSinha, D. K., Nagaraju, J., Tomar, A., Bentur, J. S., & Nair, S. (2012). Pyrosequencing-Based Transcriptome Analysis of the Asian Rice Gall Midge Reveals Differential Response during Compatible and Incompatible Interaction. International Journal of Molecular Sciences, 13(10), 13079-13103. https://doi.org/10.3390/ijms131013079





