Molecular Cloning, Characterization and Predicted Structure of a Putative Copper-Zinc SOD from the Camel, Camelus dromedarius
Abstract
:1. Introduction
2. Results
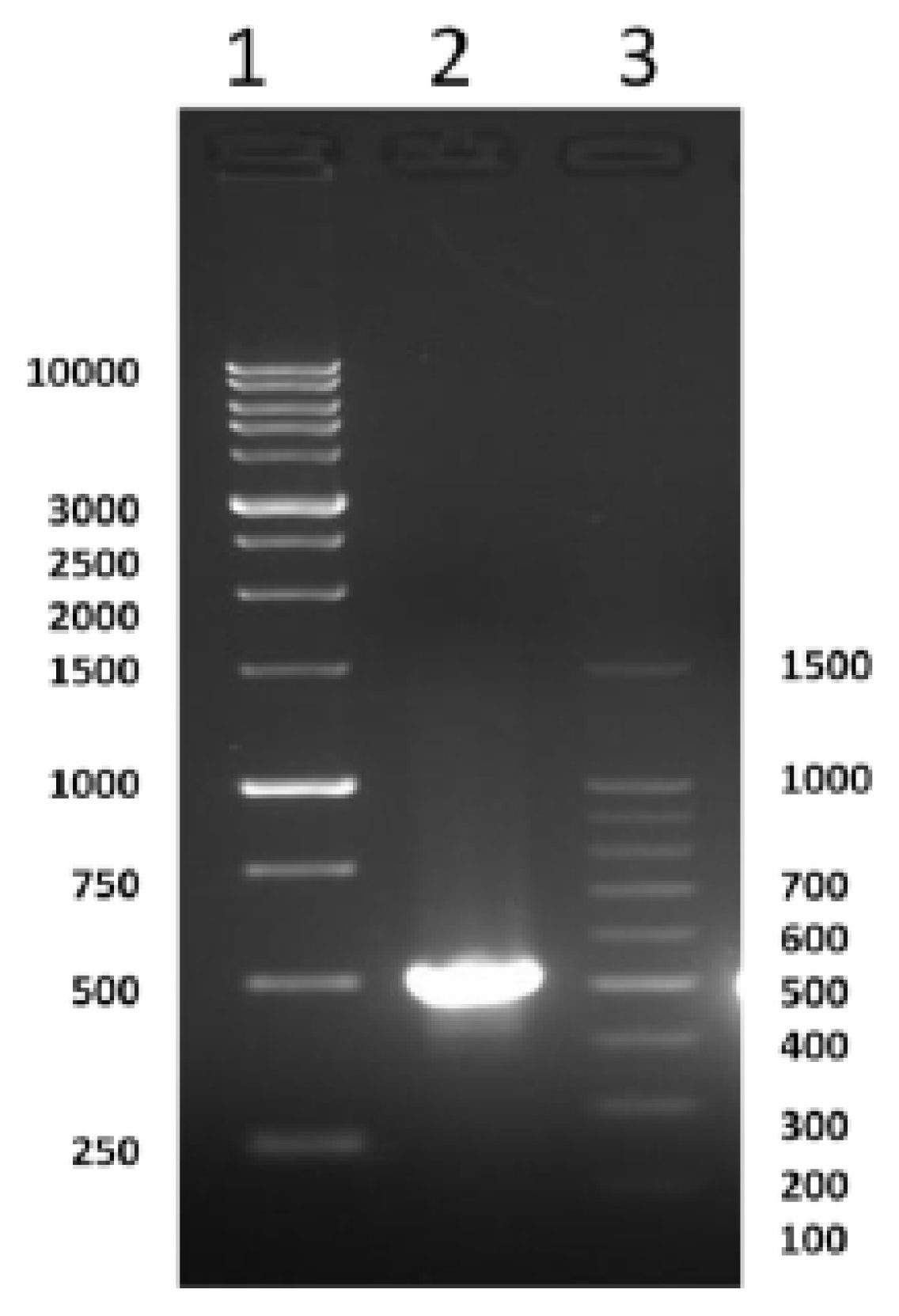
2.1. Cloning and Characterization of Full Coding of cSOD1 Gene
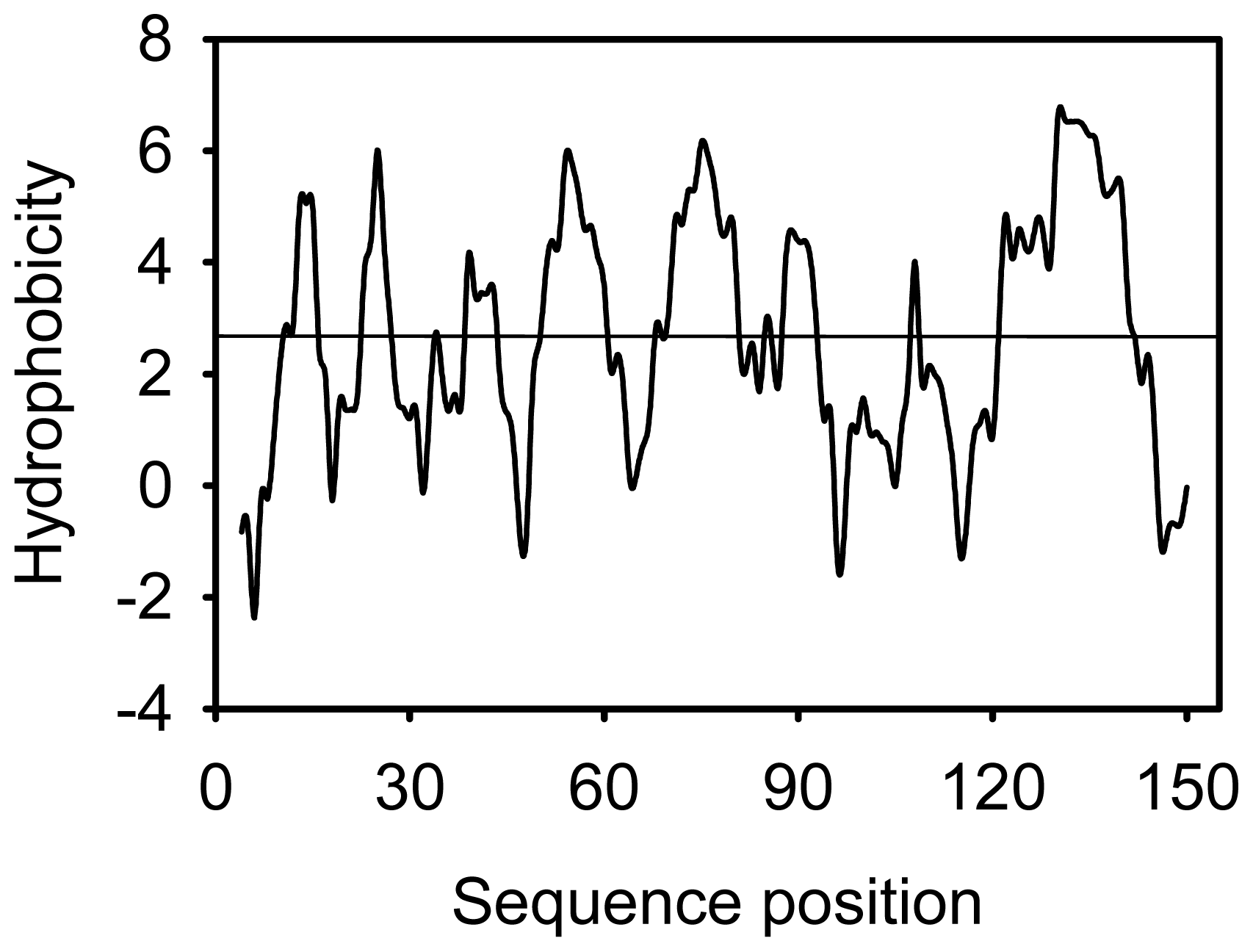
2.2. Amino Acid Composition and Protein Secondary Structure
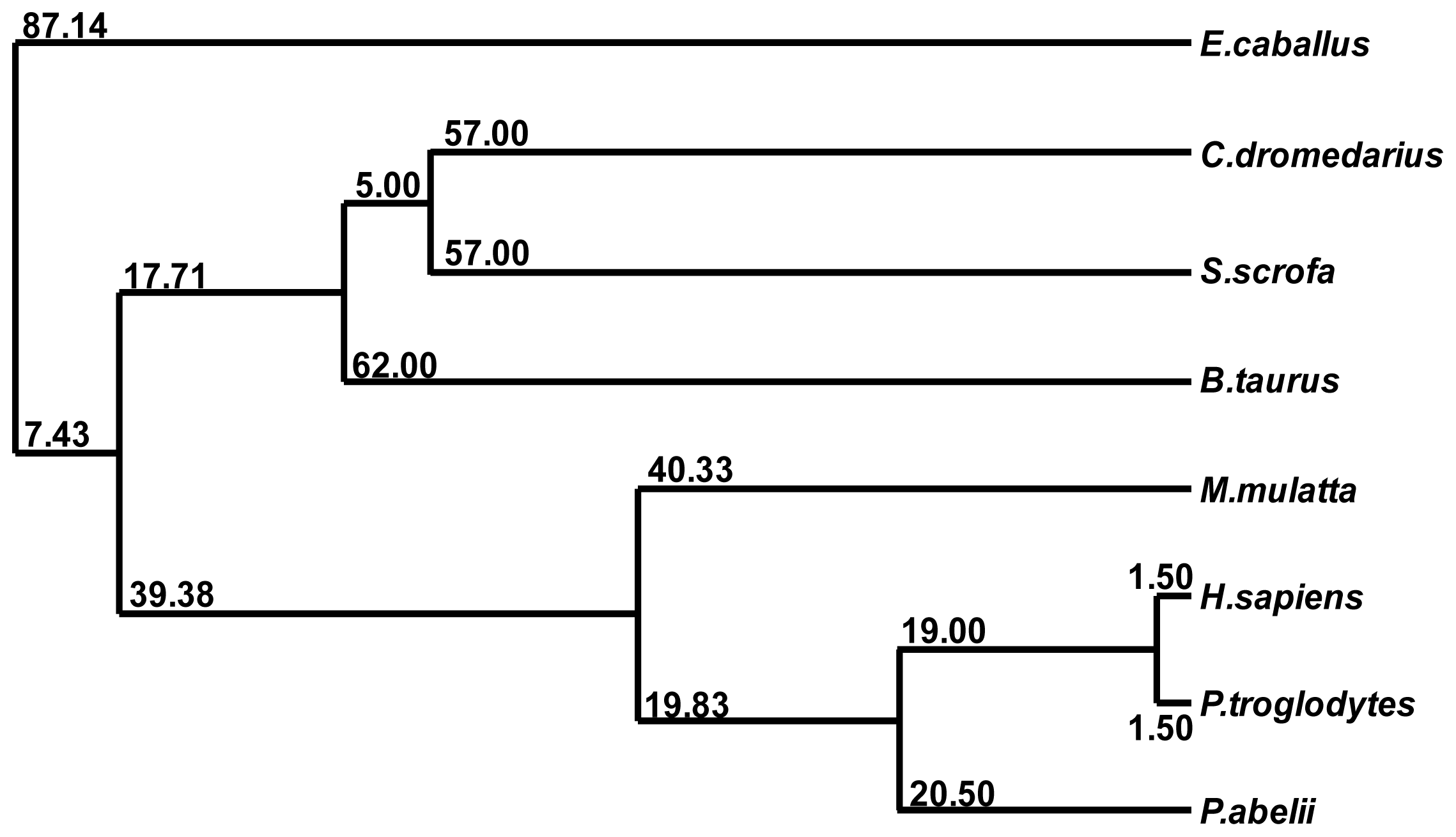
2.3. Multiple Sequence Alignment
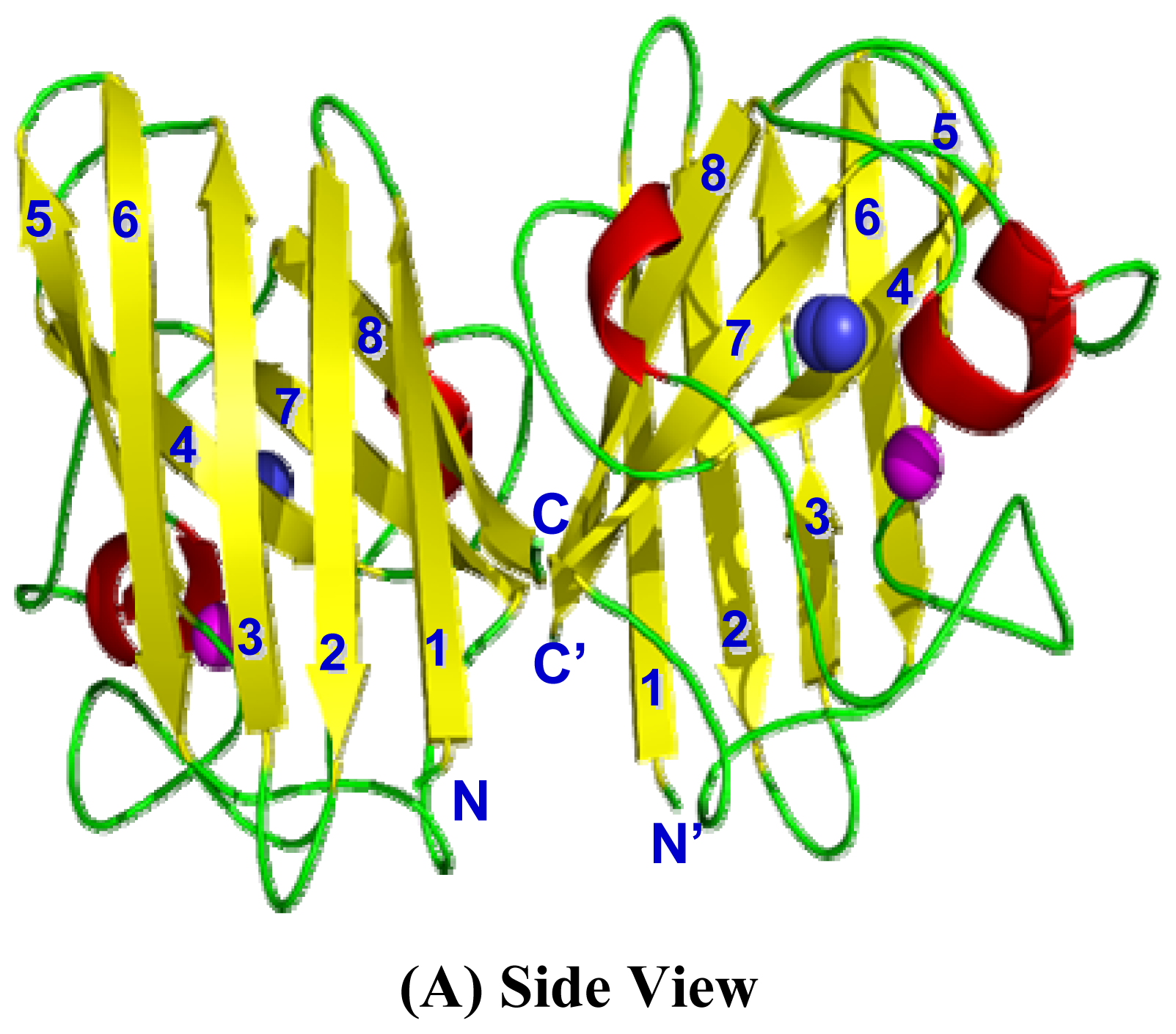
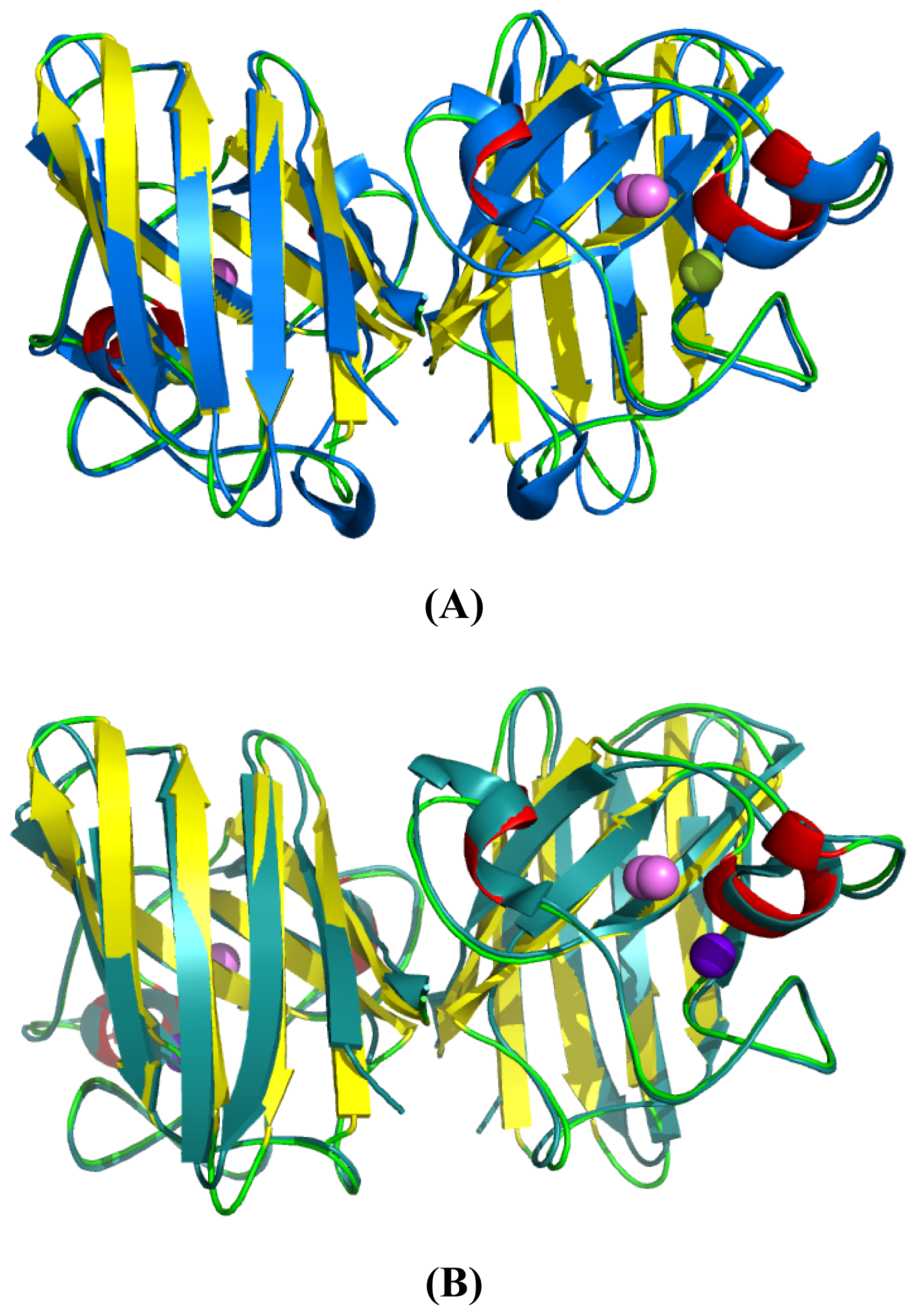
2.4. 3D Structure Prediction
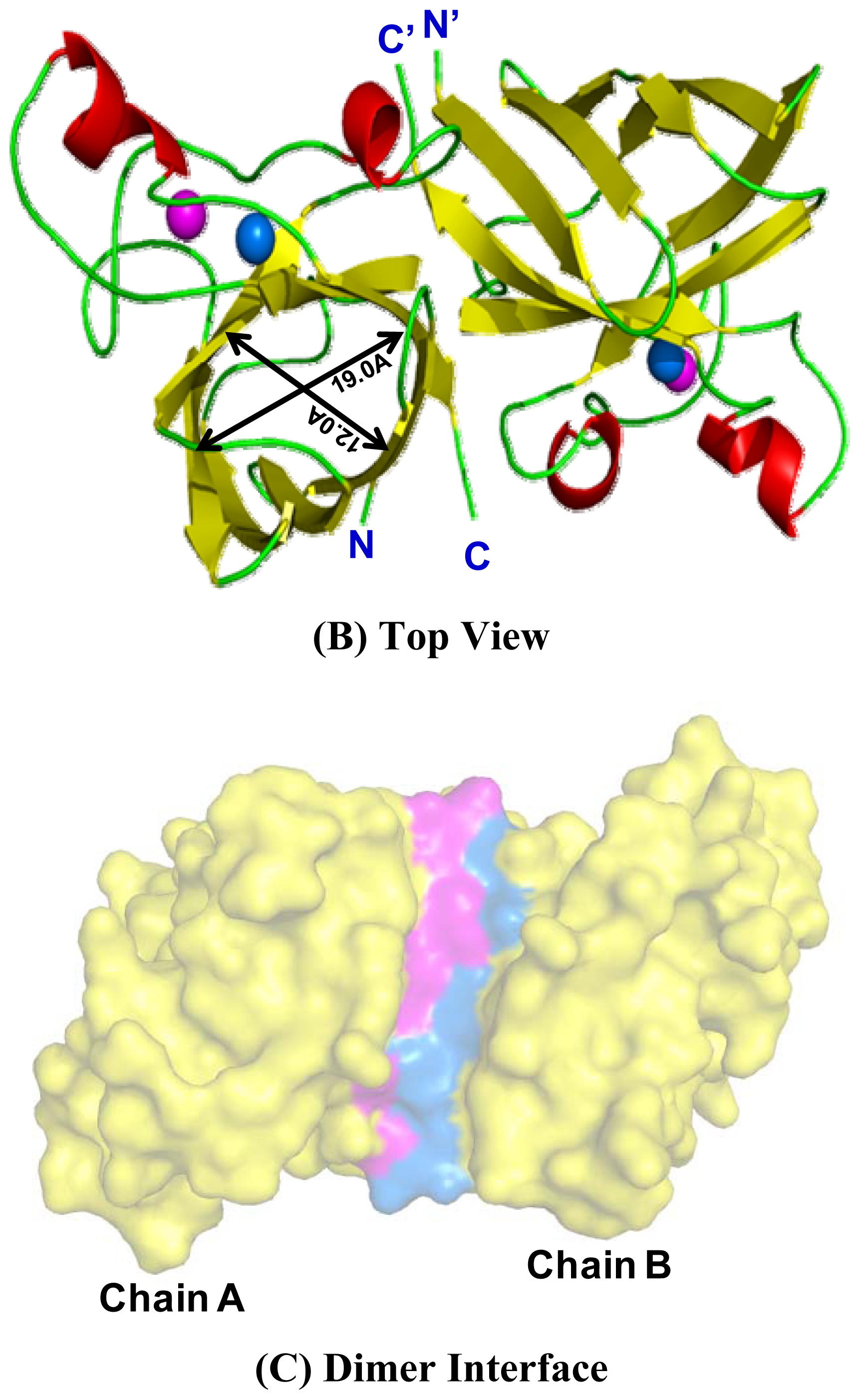
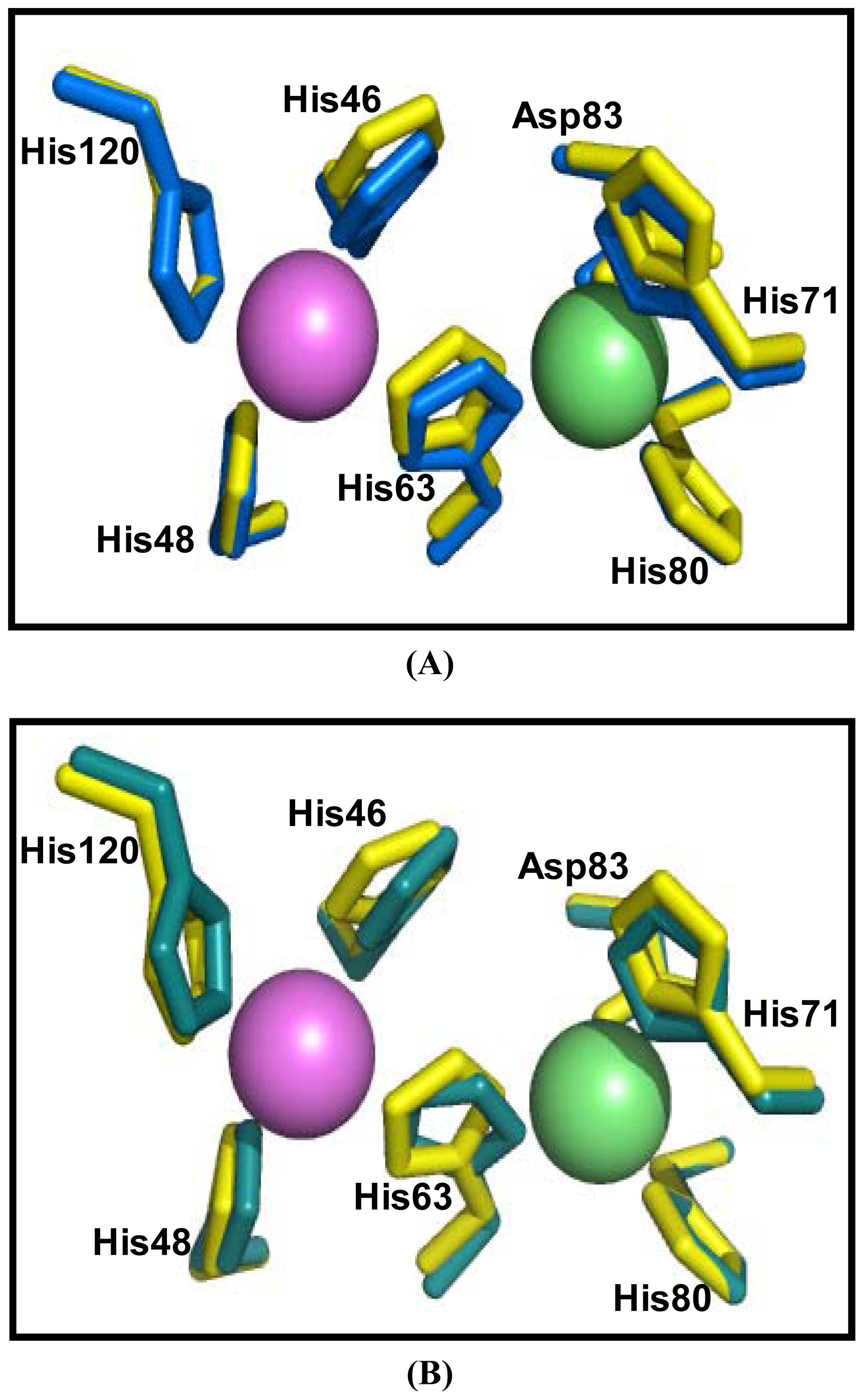
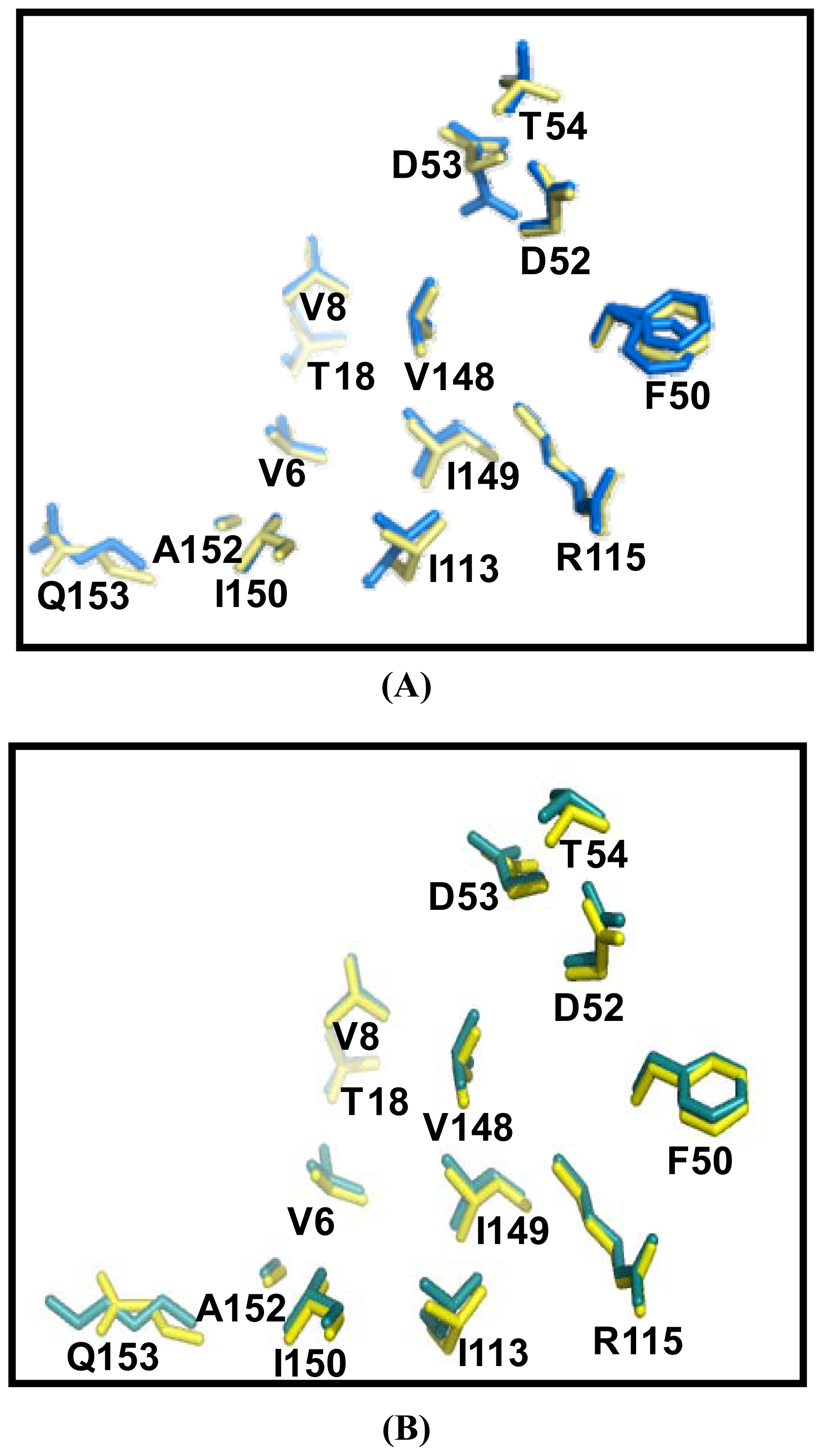
2.5. Similarities Between Structure of Camel SOD1 and Other Mammalian SOD1
2.6. Expression of cSOD1 Gene by Real Time PCR
3. Discussion
4. Experimental Section
4.1. Samples and Materials
4.2. Oligonucleotide Design
4.3. RNA Extraction, cDNA Synthesis and Reverse Transcription PCR
4.4. Polymerase Chain Reaction and Cloning
4.5. Studying Gene Expression by qPCR
4.6. Sequencing of the PCR Products and Prediction of Amino Acid Sequence
4.7. Multiple Sequence Alignment and Phylogenetic Analysis
4.8. Secondary and 3D Structure Prediction of cSOD1, Superimposition, Antigenicity and Hydrophilicity
Acknowledgments
References
- Redwan, R.M.; Tabll, A. Camel lactoferrin markedly inhibits hepatitis C virus genotype 4 infection of human peripheral blood leukocytes. J. Immunoass. Immunochem 2007, 28, 267–277. [Google Scholar]
- Halliwell, B. Free radicals, antioxidants and human disease: Curiosity, cause or consequence. Lancet 1994, 344, 721–724. [Google Scholar]
- Halliwell, B. Free radicals, protein and DNA: Oxidative damage versus redox regulation. Biochem. Soc. Trans 1996, 24, 1023–1027. [Google Scholar]
- Fridovich, I. The biology of oxygen radicals. Science 1978, 201, 875–880. [Google Scholar]
- Turrens, J.F. Mitochondrial formation of reactive oxygen species. J. Physiol 2003, 15, 335–344. [Google Scholar]
- Rapp, U.; Adams, W.; Miller, R. Purification of superoxide dismutase from fungi and characterization of the reaction of the enzyme with catechols by electron spin resonance spectroscopy. Can. J. Biochem 1973, 51, 158–171. [Google Scholar]
- Gregory, E.; Yost, F.; Fridovich, I. Superoxide dismutases of Escherichia coli: Intracellular localization and functions. J. Bacteriol 1973, 115, 987–991. [Google Scholar]
- Goscin, S.; Fridovich, I. The purification and properties of superoxide dismutase from Saccharomyces cerevisiae. Biochim. Biophys. Acta 1972, 289, 276–283. [Google Scholar]
- Li, H.Y.; Zhao, Y.; Wang, W.N.; Zhao, D.Q. Purification and characterization of a superoxide dismutase from Panax ginseng. Biomed. Chromatogr 2010, 24, 1203–1207. [Google Scholar]
- Aydemir, T.; Tarhan, L. Purifcation and partial characterisation of superoxide dismutase from chicken erythrocytes. Turk. J. Chem 2001, 25, 451–459. [Google Scholar]
- Karadag, H.; Bilgin, R. Purification of copper-zinc superoxide dismutase from human erythrocytes and partial characterization. Biotechnol. Biotechnol. Equip 2010, 24, 1653–1656. [Google Scholar]
- Zelko, I.N.; Mariani, T.J.; Folz, R.J. Superoxide dismutase multigene family: A comparison of the CuZnSOD (SOD1), Mn-SOD1 (SOD2) and EC-SOD1 (SOD3) gene structures, evolution, and expression. Free Radic. Biol. Med 2002, 33, 337–349. [Google Scholar]
- Fridovich, I. Superoxide dismutases. Adv. Enzymol. Relat. Areas Mol. Biol 1974, 41, 35–97. [Google Scholar]
- Lumsden, J.; Hall, D.O. Soluble and membrane-bound superoxide dismutases in a blue-green algae (Spirulina) and spinach. Biochem. Biophys. Res. Commun 1974, 58, 35–41. [Google Scholar]
- Forman, H.; Fridovich, I. On the stability of bovine superoxide dismutase. The effects of metals. J. Biol. Chem 1973, 248, 2645–2649. [Google Scholar]
- Banci, L.; Bertini, I.; Cabelli, D.E.; Hallewell, R.A.; Tung, J.W.; Viezzoli, M.S. A characterization of copper/zinc superoxide dismutase mutants at position 124. Zinc-deficient proteins. Eur. J. Biochem 1991, 196, 123–128. [Google Scholar]
- Borchelt, D.R.; Lee, M.K.; Slunt, H.S.; Guarnieri, M.; Xu, Z.S.; Wong, P.C.; Brown, R.H.; Price, D.L.; Sisodia, S.S.; Cleveland, D.W. Superoxide dismutase 1 with mutations linked to familial amyotrophic lateral sclerosis possesses significant activity. Proc. Natl. Acad. Sci. USA 1994, 91, 8292–8296. [Google Scholar]
- Fee, J.; Teitelbaum, H. Evidence that superoxide dismutase plays a role in protecting red blood cells against peroxidative hemolysis. Biochem. Biophys. Res. Commun 1972, 49, 150–158. [Google Scholar]
- Khan, M.A.; Tania, M.; Zhang, D.; Chen, H. Antioxidant enzymes and cancer. Chin. J. Cancer Res 2010, 22, 87–92. [Google Scholar]
- Deng, H.X.; Hentati, A.; Tainer, J.A.; Iqbal, Z.; Cayabyab, A.; Hung, W.Y.; Getzoff, E.D.; Hu, P.; Herzfeld, B.; Roos, R.P.; et al. Amyotrophic lateral sclerosis and structural defects in Cu, Zn superoxide dismutase. Science 1993, 261, 1047–1051. [Google Scholar]
- Brown, R.H. Amyotrophic lateral sclerosis: Recent insights from genetics and transgenic mice. Cell 1995, 80, 687–692. [Google Scholar]
- Al-Chalabi, A.; Leigh, P.N. Recent advances in amyotrophic lateral sclerosis. Curr. Opin. Neurol 2000, 13, 397–405. [Google Scholar]
- Groner, Y.; Elroy-Stein, O.; Avraham, K.B.; Schickler, M.; Knobler, H.; Minc-Golomb, D.; Bar-Peled, O.; Yarom, R.; Rotshenker, S. Cell damage by excess CuZnSOD and Down syndrome. Biomed. Pharmacother 1994, 48, 231–240. [Google Scholar]
- Celino, F.T.; Yamaguchi, S.; Miura, C.; Ohta, T.; Tozawa, Y.; Iwai, T.; Miura, T. Tolerance of spermatogonia to oxidative stress is due to high levels of Zn and Cu/Zn superoxide dismutase. PLoS One 2011, 6, e16938. [Google Scholar]
- Imamura, Y.; Noda, S.; Hashizume, K.; Shinoda, K.; Yamaguchi, M.; Uchiyama, S.; Shimizu, T.; Mizushima, Y.; Shirasawa, T.; Tsubota, K. Drusen, choroidal neovascularization, and retinal pigment epithelium dysfunction in SOD1-deficient mice: A model of age-related macular degeneration. Proc. Natl. Acad. Sci. USA 2006, 103, 11282–11287. [Google Scholar]
- Uchiyama, S.; Shimizu, T.; Shirasawa, T. CuZnSOD deficiency causes ApoB degradation and induces hepatic lipid accumulation by impaired lipoprotein secretion in mice. J. Biol. Chem 2006, 281, 31713–31719. [Google Scholar]
- Murakami, K.; Inagaki, J.; Saito, M.; Ikeda, Y.; Tsuda, C.; Noda, Y.; Kawakami, S.; Shirasawa, T.; Shimizu, T. Skin atrophy in cytoplasmic SOD-deficient mice and its complete recovery using a vitamin C derivative. Biochem. Biophys. Res. Commun 2009, 382, 457–461. [Google Scholar]
- Elchuri, S.; Oberley, T.D.; Qi, W.; Eisenstein, R.S.; Roberts, L.J.; van Remmen, H.; Epstein, C.J.; Huang, T.T. CuZnSOD deficiency leads to persistent and widespread oxidative damage and hepatocarcinogenesis later in life. Oncogene 2005, 24, 367–380. [Google Scholar]
- Iuchi, Y.; Okada, F.; Onuma, K.; Onoda, T.; Asao, H.; Kobayashi, M.; Fujii, J. Elevated oxidative stress in erythrocytes due to a SOD1 deficiency causes anaemia and triggers autoantibody production. Biochem. J 2007, 402, 219–227. [Google Scholar]
- Muller, F.L.; Song, W.; Liu, Y.; Chaudhuri, A.; Pieke-Dahl, S.; Strong, R.; Huang, T.T.; Epstein, C.J.; Roberts, L.J., 2nd; Csete, M.; et al. Absence of CuZn superoxide dismutase leads to elevated oxidative stress and acceleration of age-dependent skeletal muscle atrophy. Free Radic. Biol. Med 2006, 40, 1993–2004. [Google Scholar]
- Ho, Y.S.; Gargano, M.; Cao, J.; Bronson, R.T.; Heimler, I.; Hutz, R.J. Reduced fertility in female mice lacking copper-zinc superoxide dismutase. J. Biol. Chem 1998, 273, 7765–7769. [Google Scholar]
- Noda, Y.; Ota, K.; Shirasawa, T.; Shimizu, T. Copper/zinc superoxide dismutase insufficiency impairs progesterone secretion and fertility in female mice. Biol. Reprod 2011. [Google Scholar] [CrossRef]
- PROTEAN, version 5.07; DNASTAR, Inc.: Madison, WI, USA, 2003.
- MAFFT, version 6.864; Computational Biology Research Center (CBRC): Tokyo, Japan, 2011.
- PSIPRED, version 3.0; University College London: London, UK, 2008.
- Jalview, version 2.3; University of Dundee: Scotland, UK, 2011.
- Getzoff, E.D.; Tainer, J.A.; Stempien, M.M.; Bell, G.I.; Hallewell, R.A. Evolution of CuZn superoxide dismutase and the Greek key beta-barrel structural motif. Proteins 1989, 5, 322–336. [Google Scholar]
- Arnold, K.; Bordoli, L.; Kopp, J.; Schwede, T. The SWISS-MODEL Workspace: A web-based environment for protein structure homology modelling. Bioinformatics 2006, 22, 195–201. [Google Scholar]
- Rakhit, R.; Chakrabartty, A. Structure, folding, and misfolding of Cu, Zn superoxide dismutase in amyotrophic lateral sclerosis. Biochim. Biophys. Acta 2006, 1762, 1025–1037. [Google Scholar]
- Kiefer, F.; Arnold, K.; Künzli, M.; Bordoli, L.; Schwede, T. The SWISS-MODEL repository and associated resources. Nucleic Acids Res 2009, 37, 387–392. [Google Scholar]
- PyMOL, version 0.99; Schrödinger: New York, NY, USA, 2006.
- EMBL-EBI. Available online: http://www.ebi.ac.uk/msd-srv/ssm/cgi-bin/ssmserver accessed on 2 December 2011.
- Kolaskar, A.S.; Tongaonkar, P.C. A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett 1990, 276, 172–174. [Google Scholar]
- Parker, J.M.; Guo, D.; Hodges, R.S. New hydrophilicity scale derived from high-performance liquid chromatography peptide retention data: Correlation of predicted surface residues with antigenicity and X-ray-derived accessible sites. Biochemistry 1986, 25, 5425–5432. [Google Scholar]
- Tainer, J.A.; Getzoff, E.D.; Beem, K.M.; Richardson, J.S.; Richardson, D.C. Determination and analysis of the 2 A-structure of copper, zinc superoxide dismutase. J. Mol. Biol 1982, 160, 181–217. [Google Scholar]
- Valentine, J.S.; Doucette, P.A.; Potter, S.Z. Copper-zinc superoxide dismutase and amyotrophic lateral sclerosis. Annu. Rev. Biochem 2005, 74, 563–593. [Google Scholar]
- Hart, P.J.; Balbirnie, M.M.; Ogihara, N.L.; Nersissian, A.M.; Weiss, M.S.; Valentine, J.S.; Eisenberg, D. A structure-based mechanism for copper–zinc superoxide dismutase. Biochemistry 1999, 38, 2167–2178. [Google Scholar]
- Rakhit, R.; Chakrabartty, A. Structure, folding, and misfolding of Cu, Zn superoxide dismutase in amyotrophic lateral sclerosis. Biochim. Biophys. Acta 2006, 1762, 1025–1037. [Google Scholar]
- Getzoffm, E.D.; Tainer, J.A.; Olson, A.J. Recognition and interactions controlling the assemblies of beta barrel domains. Biophys. J 1986, 49, 191–206. [Google Scholar]
- Furukawa, Y.; O’Halloran, T.V. Amyotrophic lateral sclerosis mutations have the greatest destabilizing effect on the apo, reduced form of SOD1, leading to unfolding and oxidative aggregation. J. Biol. Chem 2005, 280, 17266–17274. [Google Scholar]
- Pace, C.N.; Grimsley, G.R.; Thomson, J.A.; Barnett, B.J. Conformational stability and activity of ribonuclease T1 with zero, one, and two intact disulfide bonds. J. Biol. Chem 1988, 263, 11820–11825. [Google Scholar]
- Krissinel, E.; Henrick, K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr 2004, 60, 2256–2268. [Google Scholar]
- Holm, L.; Kääriäinen, S.; Rosenström, P.; Schenkel, A. Searching protein structure databases with DaliLite v.3. Bioinformatics 2008, 24, 2780–2781. [Google Scholar]
- Marklund, S.L. Extracellular superoxide dismutase and other superoxide dismutase isoenzymes in tissues from nine mammalian species. Biochem. J 1984, 222, 649–655. [Google Scholar]
- Sambrook, J.; Fritsch, E.; Manaiatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar]
- Seqman, version 5.07; DNASTAR, Inc.: Madison, WI, USA, 2003.
- EditSeq, version 5.07; DNASTAR, Inc.: Madison, WI, USA, 2003.
- NCBI (National Center for Biotechnology Information). Available online: http://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastp&BLAST_PROGRAMS=blastp&PAGE_TYPE=BlastSearch&SHOW_DEFAULTS=on&LINK_LOC=blasthome accessed on 28 November 2011.





| Amino Acid | Number Count | % By Weight | % By Frequency | Amino Acid | Number Count | % By Weight | % By Frequency |
|---|---|---|---|---|---|---|---|
| A Ala | 9 | 4.06 | 5.88 | M Met | 3 | 2.50 | 1.96 |
| C Cys | 3 | 1.97 | 1.96 | N Asn | 6 | 4.35 | 3.92 |
| D Asp | 10 | 7.31 | 6.54 | P Pro | 6 | 3.70 | 3.92 |
| E Glu | 8 | 6.56 | 5.23 | Q Gln | 7 | 5.70 | 4.58 |
| F Phe | 4 | 3.74 | 2.61 | R Arg | 3 | 2.98 | 1.96 |
| G Gly | 26 | 9.42 | 16.99 | S Ser | 11 | 6.08 | 7.19 |
| H His | 9 | 7.84 | 5.88 | T Thr | 7 | 4.50 | 4.58 |
| I Ile | 9 | 6.47 | 5.88 | V Val | 14 | 8.82 | 9.15 |
| K Lys | 10 | 8.14 | 6.54 | W Trp | 0 | 0.00 | 0.00 |
| L Leu | 8 | 5.75 | 5.23 | Y Tyr | 0 | 0.00 | 0.00 |
| SOD | (NCBI Reference Sequence) | No. of Residues | Total Score | Coverage (%) | Identity (%) | Positive (%) | Gap (%) | E-Value | pI | MW |
|---|---|---|---|---|---|---|---|---|---|---|
| Arabian Camel, Camelus dromedarius | AEF32527 | 153 | 307 | 100 | 100 | 100 | 0 | 6.00E-89 | 5.94 | 15.70 |
| Rhesus Monkey, Macaca mulatta | NP_001027976.1 | 154 | 265 | 100 | 87 | 92 | 1 | 2.00E-76 | 6.22 | 15.90 |
| Cattle, Bos taurus | XP_584414.4 | 152 | 263 | 100 | 86 | 93 | 1 | 6.00E-76 | 5.85 | 15.60 |
| Human, Homo sapiens | NP_000445.1 | 154 | 261 | 100 | 87 | 90 | 1 | 3.00E-75 | 5.70 | 15.90 |
| Pig, Sus scrofa | NP_001177351.1 | 153 | 270 | 100 | 88 | 92 | 0 | 7.00E-78 | 6.32 | 15.80 |
| Horse, Equus caballus | NP_001075295.1 | 154 | 246 | 99 | 82 | 89 | 1 | 1.00E-70 | 6.03 | 16.00 |
| Sumatran orangutan, Pongo abelii | NP_001125441.1 | 155 | 252 | 100 | 85 | 88 | 1 | 2.00E-72 | 5.87 | 16.10 |
| Chimpanzee, Pan troglodytes | NP_001009025.1 | 154 | 261 | 100 | 87 | 90 | 1 | 3.00E-75 | 5.70 | 15.90 |
| Query Structure | Target Structure (PDB) | No. of Residues | Aligned Residues | RMSD | Q-Score | P-Score | Z-Score |
|---|---|---|---|---|---|---|---|
| Predicted cSOD | hSOD1 (2V0A) | 152 | 151 | 0.557 | 0.948 | 24.39 | 14.64 |
| Predicted cSOD | bSOD1 (1E9O) | 152 | 150 | 0.425 | 0.961 | 19.95 | 13.29 |
| No. | Start Position | End Position | Peptide | Peptide Length |
|---|---|---|---|---|
| 1 | 4 | 12 | KAVCVLKGD | 9 |
| 2 | 28 | 38 | PVMVSGSISGL | 11 |
| 3 | 44 | 50 | GFHVHQF | 7 |
| 4 | 63 | 69 | HFNPLSK | 7 |
| 5 | 92 | 113 | GDVAIVSIEDPVISLSGDHSII | 22 |
| 6 | 115 | 122 | RTMVVHEK | 8 |
© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Ataya, F.S.; Fouad, D.; Al-Olayan, E.; Malik, A. Molecular Cloning, Characterization and Predicted Structure of a Putative Copper-Zinc SOD from the Camel, Camelus dromedarius. Int. J. Mol. Sci. 2012, 13, 879-900. https://doi.org/10.3390/ijms13010879
Ataya FS, Fouad D, Al-Olayan E, Malik A. Molecular Cloning, Characterization and Predicted Structure of a Putative Copper-Zinc SOD from the Camel, Camelus dromedarius. International Journal of Molecular Sciences. 2012; 13(1):879-900. https://doi.org/10.3390/ijms13010879
Chicago/Turabian StyleAtaya, Farid S., Dalia Fouad, Ebtsam Al-Olayan, and Ajamaluddin Malik. 2012. "Molecular Cloning, Characterization and Predicted Structure of a Putative Copper-Zinc SOD from the Camel, Camelus dromedarius" International Journal of Molecular Sciences 13, no. 1: 879-900. https://doi.org/10.3390/ijms13010879




