Is Remusatia (Araceae) Monophyletic? Evidence from Three Plastid Regions
Abstract
:1. Introduction
2. Results and Discussion
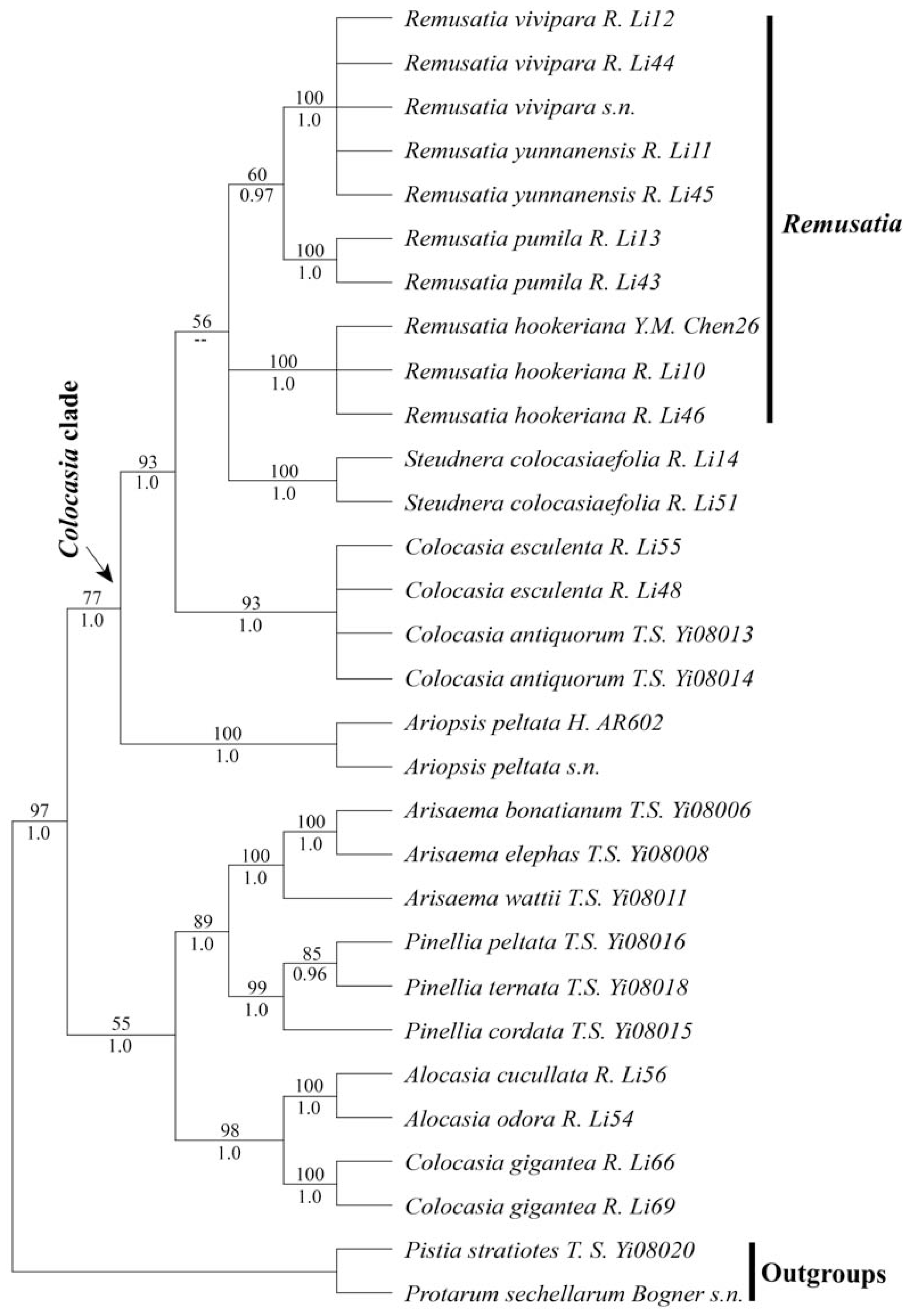
2.1. Results
2.2. Discussion
2.2.1. Is Remusatia Monophyletic?
2.2.2. Infrageneric Relationships Within Remusatia
| 1a. Spathe with two constrictions, one separating tube and limb, one separating limb into two parts | R. pumila |
| 1b. Spathe with only one constriction separating tube and limb | |
| 2a. Stonlons erect, simple, stout | R. vivipara |
| 2b. Stonlons creeping or pendulous, simple or branched, slender | |
| 3a. Limb of spathe semispreading to erect, not reflexed | R. hookeriana |
| 3b. Limb of spathe initially erect, later reflexed | R. yunnanensis |
3. Experimental Section
3.1. Taxon Sampling
3.2. DNA Extractions, Amplification, and Sequencing
3.3. Sequence Alignment and Phylogenetic Analyses
4. Conclusions
Supplementary Materials
ijms-13-00071-s001.pdfAcknowledgments
References
- Mayo, S.J.; Bogner, J.; Boyce, P.C. The Genera of Araceae; Royal Botanic Gardens: Kew, UK, 1997; pp. 280–281. [Google Scholar]
- Frodin, D.G.; Govaerts, R. World Checklist and Bibliography of Araceae; Royal Botanic Gardens: Kew, UK, 2002; pp. 440–442. [Google Scholar]
- Mabberley, D.J. Mabberley’s Plant-Book: A Portable Dictionary of Plants, Their Classification and Uses; Cambridge University Press: New York, USA, 2008; p. 731. [Google Scholar]
- Li, H.; Boyce, P.C. Wu, Z.Y., Raven, P.H., Eds.; Remusatia. In Flora of China; Science Press: Beijing, China; Missouri Botanical Garden Press: St. Louis, MO, USA, 2010; Volume 23, pp. 71–72. [Google Scholar]
- Li, H.; Hay, A. Notes on the classification of genera Remusatia and Gonatanthus in Araceae. 1992, 27–33. [Google Scholar]
- Engler, A.; Krause, K. Engler, A., Ed.; Additamentum ad Araceas-Philodendroideas, Araceae-Colocasioideae. In Das Pflanzenreich; Wilhelm Engelmann: Leipzig, Germany, 1920; Volume 71, p. 19. [Google Scholar]
- Li, H. Wu, Z.Y., Li, H., Eds.; Gonatanthus. In Flora Reipublicae Popularis Sinicae; Science Press: Beijing, China, 1979; Volume 13, pp. 62–65. [Google Scholar]
- Li, H.; Hay, A. Classification of the genus Gonatanthus. Acta Bot. Yunnanica 1992, 14, 373–378. [Google Scholar]
- Cusimano, N.; Bogner, J.; Mayo, S.J.; Boyce, P.C.; Wong, S.Y.; Hesse, M.; Hetterscheid, W.L.A.; Keating, R.C.; French, J.C. Relationships within the Araceae: Comparison of morphological patterns with molecular phylogenies. Am. J. Bot 2011, 98, 654–668. [Google Scholar]
- Renner, S.S.; Zhang, L.B. Biogeography of the Pistia clade (Araceae): Based on chloroplast and mitochondrial DNA sequences and Bayesian divergence time inference. Syst. Biol 2004, 53, 422–432. [Google Scholar]
- Cabrera, L.I.; Salazar, G.A.; Chase, M.W.; Mayo, S.J.; Bogner, J.; Davila, P. Phylogenetic relationships of aroids and duckweeds (Araceae) inferred from coding and noncoding plastid DNA. Am. J. Bot 2008, 95, 1153–1165. [Google Scholar]
- Rothwell, G.W.; van Atta, M.R.; Ballard, H.W., Jr; Stockey, R.A. Molecular phylogenetic relationships among Lemnaceae and Araceae using the chloroplast trnL-trnF spacer. Mol. Phylogenet. Evol 2004, 30, 378–385. [Google Scholar]
- Tam, S.M.; Boyce, P.C.; Upson, T.M.; Barabé, D.; Bruneau, A.; Forest, F.; Parker, J.S. Intergeneric and infrafamilial phylogeny of subfamily Monsteroideae (Araceae) revealed by chloroplast trnL-F sequences. Am. J. Bot 2004, 91, 490–498. [Google Scholar]
- Nie, Z.L.; Sun, H.; Li, H.; Wen, J. Intercontinental biogeography of subfamily Orontioideae (Symplocarpus, Lysichiton, and Orontium) of Araceae in eastern Asia and North America. Mol. Phylogenet. Evol 2006, 40, 155–165. [Google Scholar]
- Mansion, G.; Rosenbaum, G.; Schoenenberger, N.; Bacchetta, G.; Rosselló, J.A.; Conti, E. Phylogenetic analysis informed by geological history supports multiple, sequential invasions of the Mediterranean Basin by the angiosperm family Araceae. Syst. Biol 2008, 57, 269–285. [Google Scholar]
- Wong, S.Y.; Boyce, P.C.; Othman, A.S.; Leaw, C.P. Molecular phylogeny of tribe Schismatoglottideae (Araceae) based on two plastid markers and recognition of a new tribe, Philonotieae, from the Neotropics. Taxon 2010, 59, 117–124. [Google Scholar]
- Espíndola, A.; Buerki, S.; Bedalov, M.; Kupfer, P.; Alvarez, N. New insights into the phylogenetics and biogeography of Arum (Araceae): Unravelling its evolutionary history. Bot. J. Linn. Soc 2010, 163, 14–32. [Google Scholar]
- Li, H. What is Gonatanthus (?) ornatus Schott (Araceae). Aroideana 1987, 10, 23–26. [Google Scholar]
- Li, H. A new combination of the genus Remusatia of Araceae. Acta Phytotaxon. Sin 1987, 25, 414–416. [Google Scholar]
- Li, H. On the typification of two species in the genus Remusatia (Araceae). Acta Bot. Yunnanica 1991, 13, 113–119. [Google Scholar]
- Li, H. Formation of distribution area of Remusatia (Araceae) and its disjunction. 1992, 71–76. [Google Scholar]
- Long, C.L.; Li, H.; Liu, X.Z.; Gu, Z.J. A cytogeographic study of the genus Remusatia (Araceae). Acta Bot. Yunnanica 1989, 11, 132–138. [Google Scholar]
- Mahabalé, T.S.; Deshpande, G.S. Bulbils of Remusatia vivipara Schott. J. Univ. Bombay 1938, 6, 47–56. [Google Scholar]
- Doyle, J.J.; Doyle, J.L. A rapid isolation procedure from small quantities of fresh leaf tissue. Phytochem. Bull 1987, 19, 11–15. [Google Scholar]
- Asmussen, C.B.; Chase, M.W. Coding and noncoding plastid DNA in palm systematics. Am. J. Bot 2001, 88, 1103–1117. [Google Scholar]
- Taberlet, P.; Gielly, L.; Pautou, G.; Bouvet, J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol. Biol 1991, 17, 1105–1109. [Google Scholar]
- Bellstedt, D.U.; Linder, H.P.; Harley, E. Phylogenetic relationships in Disa based on non-coding trnL-trnF chloroplast sequences: Evidence of numerous repeat regions. Am. J. Bot 2001, 88, 2088–2100. [Google Scholar]
- Oxelman, B.; Liden, M.; Berglund, D. Chloroplast rps16 intron phylogeny of the tribe Sileneae (Caryophyllaceae). Plant Syst. Evol 1997, 206, 393–410. [Google Scholar]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989. [Google Scholar]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The Clustalx windows interface. Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 1997, 25, 4876–4882. [Google Scholar]
- Rambaut, A. Se-Al version 2.0a11. 2007. Available online: http://tree.bio.ed.ac.uk/software/seal/ (accessed on 21 December 2011).
- Farris, J.S.; Källersjö, M.; Kluge, A.; Bult, G.C. Constructing a significance test for incongruence. Syst. Biol 1995, 44, 570–572. [Google Scholar]
- Swofford, D.L. PAUP*: Phylogenetic Analysis Using Parsimony (* and Other Methods), Version 4.0b10; Sinauer Associates: Sunderland, MA, USA, 2002. [Google Scholar]
- Cunningham, C.W. Can three incongruence tests predict when data should be combined? Mol. Biol. Evol 1997, 14, 733–740. [Google Scholar]
- Simmons, M.P.; Ochoterena, H. Gaps as characters in sequence based phylogenetic analyses. Syst. Biol 2000, 49, 362–381. [Google Scholar]
- Young, N.D.; Healy, J. GapCoder automates the use of indel characters in phylogenetic analysis. BMC Bioinforma 2003, 4, 1–6. [Google Scholar]
- Felsenstein, J. Confidence intervals on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar]
- Posada, D.; Crandall, K.A. Modeltest, testing the model of DNA substitution. Bioinformatics 1998, 14, 817–818. [Google Scholar]
- Posada, D.; Buckley, T.R. Model selection and model averaging in phylogenetics, advantages of the AIC and Bayesian approaches over likelihood ratio tests. Syst. Biol 2004, 53, 793–808. [Google Scholar]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar]
- Rambaut, A.; Drummond, A.J. Tracer version 1.5. 2007. Available online: http://beast.bio.ed.ac.uk/Tracer (accessed on 21 December 2011).
- Shimodaira, H.; Hasegawa, M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol. Biol. Evol 1999, 16, 1114–1116. [Google Scholar]
| Aligned length (bp) | Number of variable sites (%) | Number of parsimony-informative sites (%) | Model selected by AIC | |
|---|---|---|---|---|
| rbcL gene | 1221 | 80 (6.6%) | 35 (2.9%) | HKY + I + G |
| trnL-F intergenic spacer | 936 | 85 (9.1%) | 45 (4.8%) | TVM + G |
| rps16 intron | 1014 | 160 (15.8%) | 73 (7.2%) | TIM + G |
| Plastid concatenated | 3171 | 325 (10.2%) | 153 (4.8%) |
| Taxon | Voucher | Locality | GenBank Accession No. | ||
|---|---|---|---|---|---|
| rbcL | trnL-F | rps16 | |||
| Ingroups | |||||
| Alocasia cucullata (Loureiro) G. Don | R. Li 56 | China: Yunnan, Xishuangbanna | JQ237188 | JQ237218 | JQ237158 |
| Alocasia odora (Roxburgh) K. Koch | R. Li 54 | China: Yunnan, Xishuangbanna | JQ237190 | JQ237220 | JQ237160 |
| Ariopsis peltata Nimmo | H. AR 602 | S India | JQ237191 | JQ237221 | JQ237161 |
| Ariopsis peltata Nimmo | s.n. | S India | JQ237192 | JQ237222 | JQ237162 |
| Arisaema bonatianum Engler | T.S. Yi 08006 | China: Yunnan, Gongshan | JQ237193 | JQ237223 | JQ237163 |
| Arisaema elephas Buchet | T.S. Yi 08008 | China: Yunnan, Baoshan | JQ237194 | JQ237224 | JQ237164 |
| Arisaema wattii J. D. Hooker | T.S. Yi 08011 | China: Yunnan, Gongshan | JQ237195 | JQ237225 | JQ237165 |
| Colocasia antiquorum Schott | T.S. Yi 08013 | China: Yunnan, Tengchong | JQ237199 | JQ237229 | JQ237169 |
| Colocasia antiquorum Schott | T.S. Yi 08014 | China: Yunnan, Yingjiang | JQ237200 | JQ237230 | JQ237170 |
| Colocasia esculenta (L.) Schott | R. Li 55 | China: Yunnan, Xishuangbanna, cultivated | JQ237189 | JQ237219 | JQ237159 |
| Colocasia esculenta (L.) Schott | R. Li 48 | China: Yunnan, Lvchun, cultivated | JQ237196 | JQ237226 | JQ237166 |
| Colocasia gigantea (Blume) J. D. Hooker | R. Li 66 | China: Yunnan, Lvchun | JQ237197 | JQ237227 | JQ237167 |
| Colocasia gigantea (Blume) J. D. Hooker | R. Li 69 | China: Yunnan, Jinping | JQ237198 | JQ237228 | JQ237168 |
| Pinellia cordata N. E. Brown | T.S. Yi 08015 | China: Fujian, Wuyishan | JQ237201 | JQ237231 | JQ237171 |
| Pinellia peltata C. Pei | T.S. Yi 08016 | China: Zhejiang, Wenzhou | JQ237202 | JQ237232 | JQ237172 |
| Ingroups | |||||
| Pinellia ternata (Thunb.) Tenore ex Breitenbach | T.S. Yi 08018 | China: Yunnan, Kunming | JQ237203 | JQ237233 | JQ237173 |
| Remusatia hookeriana Schott | Y.M. Chen 26 | China: Yunnan, Wuding | JQ237206 | JQ237236 | JQ237176 |
| Remusatia hookeriana Schott | R. Li 10 | China: Yunnan, Jinping | JQ237207 | JQ237237 | JQ237177 |
| Remusatia hookeriana Schott | R. Li 46 | China: Yunnan, Lvchun | JQ237208 | JQ237238 | JQ237178 |
| Remusatia pumila (D. Don) H. Li & A. Hay | R. Li 13 | China: Yunnan, Pingbian | JQ237209 | JQ237239 | JQ237179 |
| Remusatia pumila (D. Don) H. Li & A. Hay | R. Li 43 | China: Yunnan, Lvchun | JQ237210 | JQ237240 | JQ237180 |
| Remusatia vivipara (Roxb.) Schott | R. Li 12 | China: Yunnan, Jinping | JQ237211 | JQ237241 | JQ237181 |
| Remusatia vivipara (Roxb.) Schott | R. Li 44 | China: Yunnan, Lvchun | JQ237212 | JQ237242 | JQ237182 |
| Remusatia vivipara (Roxb.) Schott | s.n. | S India | JQ237213 | JQ237243 | JQ237183 |
| Remusatia yunnanensis (H. Li & A. Hay) H. Li & A. Hay | R. Li 11 | China: Yunnan, Yingjiang | JQ237214 | JQ237244 | JQ237184 |
| Remusatia yunnanensis (H. Li & A. Hay) H. Li & A. Hay | R. Li 45 | China: Yunnan, Yingjiang | JQ237215 | JQ237245 | JQ237185 |
| Steudnera colocasiifolia K. Koch | R. Li 14 | China: Yunnan, Puer | JQ237216 | JQ237246 | JQ237186 |
| Steudnera colocasiifolia K. Koch | R. Li 51 | China: Yunnan, Xishuangbanna | JQ237217 | JQ237247 | JQ237187 |
| Outgroups | |||||
| Pistia stratiotes L. | T.S. Yi 08020 | China: Yunnan, Kunming | JQ237204 | JQ237234 | JQ237174 |
| Protarum sechellarum Engl. | Bogner s.n. | Germany: Munich Botanical Garden, cultivated | JQ237205 | JQ237235 | JQ237175 |
© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Li, R.; Yi, T.; Li, H. Is Remusatia (Araceae) Monophyletic? Evidence from Three Plastid Regions. Int. J. Mol. Sci. 2012, 13, 71-83. https://doi.org/10.3390/ijms13010071
Li R, Yi T, Li H. Is Remusatia (Araceae) Monophyletic? Evidence from Three Plastid Regions. International Journal of Molecular Sciences. 2012; 13(1):71-83. https://doi.org/10.3390/ijms13010071
Chicago/Turabian StyleLi, Rong, Tingshuang Yi, and Heng Li. 2012. "Is Remusatia (Araceae) Monophyletic? Evidence from Three Plastid Regions" International Journal of Molecular Sciences 13, no. 1: 71-83. https://doi.org/10.3390/ijms13010071




