New User-Friendly Approach to Obtain an Eisenberg Plot and Its Use as a Practical Tool in Protein Sequence Analysis
Abstract
:1. Introduction
2. Results and Discussion
2.1. The Eisenberg Plot Approach Using the Original Databases
2.2. The Validity Check of the “New” Eisenberg Plot
2.3. The Meaning of the Eisenberg Plot for Novel Classes of Proteins and Peptides
2.4. Examples Illustrating the Power of the Total Approach
3. Method Section
3.1. Primary and Secondary Structures Identification
3.2. Determination Lipid-Binding Potential
3.3. Eisenberg Plot Approach
4. Conclusions
Supplementary Information
ijms-12-05577-s001.pdfReferences
- Eisenberg, D; Weiss, RM; Terwilliger, TC. The helical hydrophobic moment: A measure of the amphiphilicity of a helix. Nature 1982, 299, 371–374. [Google Scholar]
- Eisenberg, D; Schwarz, E; Komaromy, M; Wall, R. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J Mol Biol 1984, 15, 125–142. [Google Scholar]
- Eisenberg, D; Weiss, RM; Terwilliger, TC. The hydrophobic moment detects periodicity in protein hydrophobicity. Proc Natl Acad Sci USA 1984, 81, 140–144. [Google Scholar]
- Eisenberg, D. Three-dimensional structure of membrane and surface proteins. Annu Rev Biochem 1984, 53, 595–623. [Google Scholar]
- Phoenix, DA; Harris, F. The hydrophobic moment and its use in the classification of amphiphilic structures (review). Mol Membr Biol 2002, 19, 1–10. [Google Scholar]
- Gautier, R; Douguet, D; Anthonny, B; Drin, G. Heliquest: A web-server to screen sequences with specificα–helical properties. Bioinformatics 2008, 24, 2101–2102. [Google Scholar]
- Scott, DL; Diez, G; Goldmann, WH. Protein-lipid interactions: Correlation of a predictive algorithm for lipid-binding sites with three-dimensional structural data. Theor Biol Med Model 2006, 3. [Google Scholar] [CrossRef]
- Brzeska, H; Guag, J; Remmert, K; Chacko, S; Korn, ED. An experimentally based computer search identifies unstructured membrane-binding sites in proteins: Application to class I myosins, PAKS, and CARMIL. J Biol Chem 2010, 285, 5738–5747. [Google Scholar]
- Cserhati, T. Interaction of phospholipids with proteins and peptides. New advances III. Int J Biochem 1993, 25, 123–131. [Google Scholar]
- Dowhan, W. Molecular basis for membrane phospholipid diversity: Why are there so many lipids? Annu Rev Biochem 1997, 66, 199–232. [Google Scholar]
- Sudhahar, CG; Haney, RM; Xue, Y; Stahelin, RV. Cellular membranes and lipid-binding domains as attractive targets for drug development. Curr Drug Targets 2008, 9, 603–613. [Google Scholar]
- Keller, RCA. The prediction of novel multiple lipid-binding regions in protein translocation motor proteins: A possible general feature. Cell Mol Biol Lett 2011, 16, 40–54. [Google Scholar]
- Ulbrandt, ND; London, EL; Oliver, DB. Deep penetration of a portion of Escherichia coli SecA protein into model membranes is promoted by anionic phospholipids and by partial unfolding. J Biol Chem 1992, 267, 15184–15192. [Google Scholar]
- Breukink, E; Keller, RCA; de Kruijff, B. Nucleotide and negatively charged lipid-dependent vesicle aggregation caused by SecA. FEBS Lett 1993, 331, 19–24. [Google Scholar]
- Ahn, T; Kim, H. SecA of Escherichia coli traverses lipid bilayer of phospholipid vesicles. Biochem Biophys Res Commun 1994, 203, 326–330. [Google Scholar]
- Fauchere, J; Pliska, V. Hydrophobic parameters p of amino-acid side chains from the partitioning of N-acetyl-amino-acid amides. Eur J Med Chem 1983, 8, 369–375. [Google Scholar]
- Derossi, D; Chassaing, G; Prochiantz, A. Trojan peptides: The penetratin system for intracellular delivery. Trends Cell Biol 1998, 8, 84–87. [Google Scholar]
- Sivakamasundari, C; Nagaraj, R. Interaction of 18-residue peptides derived from amphipathic helical segments of globular proteins with model membranes. J Biosci 2009, 34, 239–250. [Google Scholar]
- Kim, YS; Cha, HJ. Disperse distribution of cationic amino acids on hydrophilic surface of helical wheel enhances antimicrobial peptide activity. Biotechn Bioeng 2010, 107, 216–223. [Google Scholar]
- Sitaram, N; Subbalakshmi, C; Nagaraj, R. Identification of a second membrane-active 13-residue peptide segment in the antimicrobial protein, bovine seminalplasmin. FEBS Lett 1993, 328, 239–242. [Google Scholar]
- Helmerhors, EJ; van’t Hof, W; Breeuweri, P; Veerman, ECI; Abee, T; Troxler, RF; Nieuw Amerongen, AV; Oppenheim, FG. Characterization of Histatin 5 with respect to amphipathicity, hydrophobicity, and effects on cell and mitochondrial membrane integrity excludes a candidacidal mechanism of pore formation. J Biol Chem 2001, 276, 5643–5649. [Google Scholar]
- Bruni, R; Taeusch, HW; Waring, AJ. Surfactant protein B: Lipid interactions of synthetic peptides representing the amino-terminal amphipathic domain. Proc Natl Acad Sci USA 1991, 88, 7451–7455. [Google Scholar]
- Feng, L; Chan, WW; Roderick, SL; Cohen, DE. High-level expression and mutagenesis of recombinant human phosphatidylcholine transfer protein using a synthetic gene: Evidence for a C-terminal membrane binding domain. Biochemistry 2000, 39, 15399–15409. [Google Scholar]
- Belmonte, G; Menestrina, G; Perderzolli, C; Krizaj, I; Gubensek, F; Macek, P. Primary and secondary structure of a pore-forming toxin from the sea anemone, Actinia equina L., and its association with lipid vesicles. Biochim Biophys Acta 1994, 1192, 197–204. [Google Scholar]
- Gustafsson, M; Vandenbussche, G; Cursted, T; Ruysschaert, J-M; Johansson, J. The 21-residue surfactant peptide (LysLeu4)4Lys(KL4) is a transmembrane α-helix with a mixed nonpolar/polar surface. FEBS Lett 1996, 384, 185–188. [Google Scholar]
- de Planque, MRR; Kruijtzer, JAW; Liskamp, RMJ; Marsh, D; Greathouse, DV; Koeppe, RE; de Kruijff, B; Killian, JA. Different membrane anchoring positions of Tryptophan and lysine in synthetic transmembrane alpha-helical peptides. J Biol Chem 1999, 274, 20839–20846. [Google Scholar]
- Treutlein, HR; Lemmon, MA; Engelman, DM; Brunger, AT. The glycophorin A transmembrane domain dimer: Sequence-specific propensity for a right-handed supercoil of helices. Biochemistry 1992, 31, 12726–12732. [Google Scholar]
- Tatulian, SA; Tamm, LK. Secondary structure, orientation, oligomerization and lipid interactions of the transmembrane domain of Influenza Hemagglutinin. Biochemistry 2000, 39, 496–507. [Google Scholar]
- Kukol, A; Torres, J; Arkin, IT. A structure for the trimeric MHC class II-associated invariant chain transmembrane domain. J Mol Biol 2002, 320, 1109–1117. [Google Scholar]
- Phoenix, DA; Harris, F; Daman, OA; Wallace, J. The prediction of amphiphilic α-helices. Curr Prot Pept Sci 2002, 3, 201–221. [Google Scholar]
- Keller, RCA; Killian, JA; de Kruijff, B. Anionic phospholipids are essential for α-helix formation of the signal peptide of prePhoE upon interaction with phospholipid vesicles. Biochemistry 1992, 31, 1672–1677. [Google Scholar]
- Jones, JD; Gierasch, LM. Effect of charged residue substitutions on the thermodynamics of signal peptide-lipid interactions for the Escherichia coli LamB signal sequence. Biophys J 1994, 67, 1546–1561. [Google Scholar]
- Rizo, J; Blanco, FJ; Kobe, B; Bruch, MD; Gierasch, LM. Conformational behavior of Escherichia coli OmpA signal peptides in membrane mimetic environments. Biochemistry 1993, 32, 4881–4894. [Google Scholar]
- Golovastov, VV; Nesmeyanova, MA. Effect of membrane phospholipid composition and charge of the signal peptide of Escherichia coli alkaline phosphatase on efficiency of its secretion. Biochemistry (Mosc) 2003, 68, 1089–1096. [Google Scholar]
- Fidelio, GD; Austen, BM; Chapman, D; Lucy, JA. Interactions of ovalbumin and of its putative signal sequence with phospholipid monolayers. Possible importance of differing lateral stabilities in protein translocation. Biochem J 1987, 244, 295–301. [Google Scholar]
- Marcotte, I; Wegener, KL; Lam, Y-H; Chia, BCS; de Planque, MRR; Bowie, JH; Auger, M; Separovic, F. Interaction of antimicrobial peptides from Australian amphibians with lipid membranes. Chem Phys Lipids 2003, 122, 107–120. [Google Scholar]
- Ambroggio, EE; Separovic, F; Bowie, JH; Fidelio, GD; Bagatolli, LA. Direct visualization of membrane leakage induced by the antibiotic peptides: Maculatin, citropin, and aurein. Biophys J 2005, 89, 1874–1881. [Google Scholar]
- Brandenburg, K; Harris, F; Dennison, S; Seydel1, U; Phoenix, D. Domain V of m-calpain shows the potential to form an oblique-orientated α-helix, which may modulate the enzyme’s activity via interactions with anionic lipid. Eur J Biochem 2002, 269, 5414–5422. [Google Scholar]
- Dennison, SR; Morto, LH; Brandenburg, K; Harris, F; Phoenix, DA. Investigations into the ability of an oblique alpha-helical template to provide the basis for design of an antimicrobial anionic amphiphilic peptide. FEBS J 2006, 273, 3792–3803. [Google Scholar]
- Bradshaw, JP; Darkes, MJM; Harroun, TA; Katsaras, J; Epand, RM. Oblique membrane insertion of viral fusion peptide probed by neutron diffraction. Biochemistry 2000, 39, 6581–6585. [Google Scholar]
- Park, S-H; Kim, H-E; Kim, C-M; Yun, H-J; Choi, E-C; Lee, B-J. Role of praline, cysteine and a disulphide bridge in the structure and activity of the anti-microbial peptide gaegurin 5. Biochem J 2002, 368, 171–182. [Google Scholar]
- Harris, F; Brandenburg, K; Seydel, U; Phoenix, D. Investigations into the mechanisms used by the C-terminal anchors of Escherichia coli penicillin-binding proteins 4, 5, 6 and 6b for membrane interaction. Eur J Biochem 2002, 269, 5821–5829. [Google Scholar]
- Duplaa, H; Convert, O; Sautereau, AM; Tocanne, JF; Chassaing, G. Binding of substance P to monolayers and vesicles made of phosphatidylcholine and/or phosphatidylserine. Biochim Biophys Acta 1992, 1107, 12–22. [Google Scholar]
- Derossi, D; Joliot, AH; Chassaing, G; Prochiantz, A. The third helix of the Antennapedia homeodomain translocates through biological membranes. J Biol Chem 1994, 269, 10444–10450. [Google Scholar]
- Mitchell, DJ; Kim, DT; Steinman, L; Fathman, CG; Rothbard, JB. Polyarginine enters cells more efficiently than other polycationic homopolymers. J Pept Res 2000, 56, 318–325. [Google Scholar]
- Galanth, C; Abbassi, F; Lequin, O; Ayala-Sanmartin, J; Ladram, A; Nicolas, P; Amiche, M. Mechanism of antibacterial action of Dermaseptin B2: Interplay between helix-hinge-helix structure and membrane curvature strain. Biochemistry 2009, 48, 313–327. [Google Scholar]
- Hu, H-J; Holley, J; He, J; Harrison, RW; Yang, H; Tai, PC; Pan, Y. To be or not to be: Predicting soluble SecAs as membrane proteins. IEEE Trans NanoBiosci 2007, 6, 168–179. [Google Scholar]
- Cooper, DB; Smith, VF; Crane, JM; Roth, HC; Lilly, AA; Randall, LL. SecA, the motor of the secretion machine, binds diverse partners on one interactive surface. J Mol Biol 2008, 382, 74–87. [Google Scholar]
- Weiche, B; Bürk, J; Angelini, S; Schiltz, E; Thumfart, J-O; Koch, H-G. A cleavable N-terminal membrane anchor is involved in membrane binding of the Escherichia coli SRP receptor. J Mol Biol 2008, 28, 761–773. [Google Scholar]
- Parlitz, R; Eitan, A; Stjepanovic, G; Bahari, L; Bange, G; Bibi, E; Sinning, I. Escherichia coli signal recognition particle receptor FtsY contains an essential and autonomous membrane-binding amphipathic helix. J Biol Chem 2007, 44, 32176–32321. [Google Scholar]
- Jordi, W; de Kruijff, B; Marsh, D. Specificity of the interaction of amino- and carboxy-terminal fragments of the mitochondrial precursor protein apocytochrome c with negatively charged phospholipids. A spin-label electron spin resonance study. Biochemistry 1989, 28, 8998–9005. [Google Scholar]
- Szeto, TH; Rowland, SL; Rothfield, LI; King, GF. Membrane localization of MinD is mediated by a C-terminal motif that is conserved across eubacteria, archaea, and chloroplasts. Proc Natl Acad Sci USA 2002, 99, 15693–15698. [Google Scholar]
- Davidson, WS; Jonas, A; Clayton, DF; George, JM. Stabilization of α-Synuclein secondary structure upon binding to synthetic membranes. J Biol Chem 1998, 273, 9443–9449. [Google Scholar]
- Rorat, T. Plant dehydrins - Tissue location, structure and function. Cell Mol Biol Lett 2006, 11, 536–556. [Google Scholar]
- Davies, SMA; Harroun, TA; Hauβ, T; Kelly, SM; Bradshaw, JP. The membrane bound N-terminal domain of human adenosine diphosphate ribosylation factor-1 (ARF1). FEBS Lett 2003, 548, 119–124. [Google Scholar]
- Drin, G; Casella, J-F; Gautier, R; Boehmer, T; Schwartz, TU; Antonny, B. A general amphipathic α–helical motif for sensing membrane curvature. Nat Struct Mol Biol 2007, 14, 138–146. [Google Scholar]
- Thomas, AM; Harmer, SC; Khambra, T; Tinker, A. Characterization of a Binding Site for Anionic Phospholipids on KCNQ1. J Biol Chem 2011, 286, 2088–2100. [Google Scholar]
- Goldmann, WH; Teodoridis, JM; Sharma, CP; Hu, B; Isenberg, G. Fragments from Actin Binding Protein (ABP-280; Filamin) insert into reconstituted lipid layers. Biochem Biophys Res Commun 1999, 259, 108–112. [Google Scholar]
- Cornut, I; Büttner, K; Dasseux, J-L; Dufourcq, J. Application to the de novo design of ideally amphipathic Leu, Lys peptides with haemolytic activity higher than that of melittin. FEBS Lett 1994, 349, 29–33. [Google Scholar]
- Prabhakaran, M. The distribution of physical, chemical and conformational properties in signal and nascent peptides. Biochem J 1990, 269, 691–696. [Google Scholar]
- von Heijne, G. Mitochondrial targeting sequences may form amphiphilic helices. EMBO J 1986, 5, 1335–13342. [Google Scholar]
- von Heijne, G. Analysis of the distribution of charged residues in the N-terminal region of signal sequences: Implications for protein export in prokaryotic and eukaryotic cells. EMBO J 1984, 3, 2315–2318. [Google Scholar]
- von Heijne, G; Abrahmsen, L. Species-specific variation in signal peptide design. Implications for secretion in foreign hosts. FEBS Lett 1989, 244, 439–446. [Google Scholar]
- Dennison, SR; Harris, F; Phoenix, DA. Are oblique orientated α-Helices used by antimicrobial peptides for membrane invasion? Prot Pept Lett 2005, 12, 27–29. [Google Scholar]
- Gallusser, A; Kuhn, A. Initial steps in protein membrane insertion. Bacteriophage M13 procoat protein binds to the membrane surface by electrostatic interaction. EMBO J 1990, 9, 2723–2729. [Google Scholar]
- Nagler, C; Nagler, G; Kuhn, A. Cysteine residues in the transmembrane regions of M13 Procoat protein suggest that oligomeric coat Proteins assemble onto phage progeny. J Bacteriol 2007, 189, 2897–2905. [Google Scholar]
- Wells, RC; Hill, RB. The cytosolic domain of Fis1 binds and reversibly clusters lipid vesicles. PLoS One 2011, 6. [Google Scholar] [CrossRef]
- Moll, RG. Protein–protein, protein–RNA and protein–lipid interactions of signal-recognition particle components in the hyperthermoacidophilic archaeon Acidianus ambivalens. Biochem J 2003, 374, 247–254. [Google Scholar]
- Reinau, ME; Otzen, DE. Stability and structure of the membrane protein transporter Ffh is modulated by substrates and lipids. Arch Biochem Biophys 2009, 492, 48–53. [Google Scholar]
- Fannjiang, Y; Cheng, WC; Lee, SJ; Qi, B; Pevsner, J; McCaffery, JM; Hill, RB; Basañez, G; Hardwick, JM. Mitochondrial Fission proteins regulate programmed cell death in Yeast. Genes Dev 2004, 18, 2785–2797. [Google Scholar]
- Geourjon, C; Deleage, G. SOPMA: Significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput Appl Biosci 1995, 11, 681–684. [Google Scholar]
- Izard, JW; Kendall, DA. Signal peptides: Exquisitely designed transport promoters. Mol Microbiol 1994, 13, 765–773. [Google Scholar]
- Giuliani, A; Pirri, G; Nicolette, SF. Antimicrobial peptides: An overview of a promising class of therapeutics. Cent Eur J Biol 2007, 2, 1–33. [Google Scholar]
- Madani, F; Lindberg, S; Langel, Ü; Futaki, S; Gräslund, A. Mechanisms of cellular uptake of cell-penetrating peptides. J Biophys 2011, 2011, 414729. [Google Scholar]
- Burn, P. Amphitropic proteins: A new class of membrane proteins. Trends Biochem Sci 1988, 13, 79–83. [Google Scholar]
- Johnson, JE; Cornell, RB. Amphitropic proteins: Regulation by reversible membrane interactions (review). Mol Membr Biol 1999, 16, 217–235. [Google Scholar]
- Halskau, Ø; Muga, A; Martínez, A. Linking new paradigms in protein chemistry to reversible membrane-protein interactions. Curr Prot Pept Sci 2009, 10, 339–359. [Google Scholar]
- Keller, RCA; Snel, MME; de Kruijff, B; Marsh, D. SecA restricts in a nucleotide-dependent manner acyl chain mobility up to the center of a phospholipid bilayer. FEBS Lett 1995, 358, 251–254. [Google Scholar]
- Triano, I; Barrera, FN; Renart, ML; Molina, ML; Fernández-Ballester, G; Poveda, JA; Fernández, AM; Encina, JA; Ferrer-Montiel, AV; Otzen, D; et al. Occupancy of nonannular lipid binding sites on KcsA greatly increases the stability of the tetrameric protein. Biochemistry 2010, 49, 5397–5404. [Google Scholar]
- Gold, VAM; Robson, A; Baob, H; Romantsovc, T; Duong, F; Collinson, I. The action of cardiolipin on the bacterial translocon. Proc Natl Acad Sci USA 2010, 107, 10044–10049. [Google Scholar]
 ), Surface seeking (
), Surface seeking (
 ) and TransMembrane (■) segments are depicted.
) and TransMembrane (■) segments are depicted.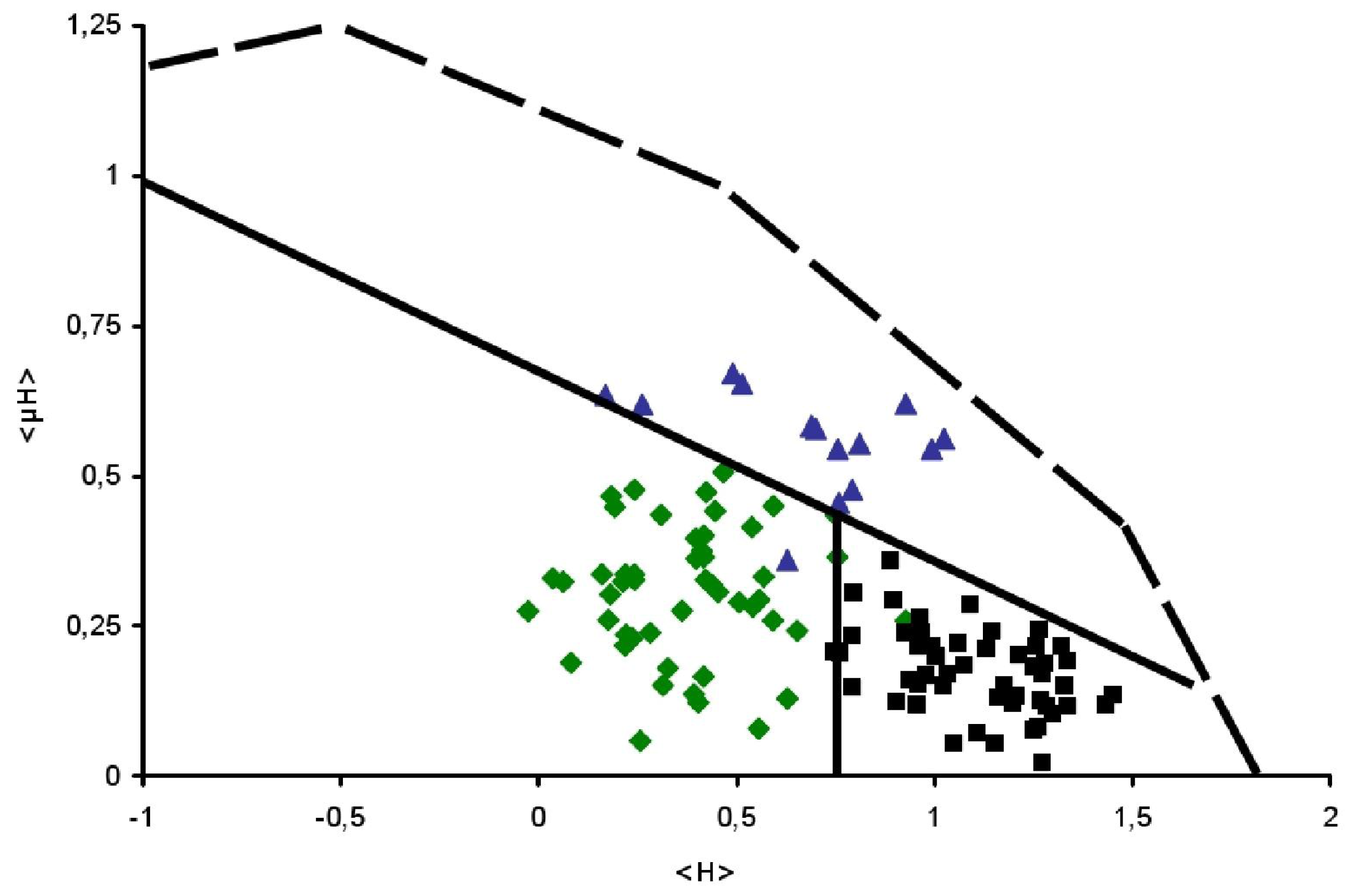
 ), Globular (
), Globular (
 ) and TransMembrane (■) segments are depicted, see Table 1 for details. Examples of signal peptides (SP) (circles, black), lipid-binding peptides (LBP) (circles, blue), amphitropics (circles, green) and others (circles, orange) are depicted, see Table 2 for details.
) and TransMembrane (■) segments are depicted, see Table 1 for details. Examples of signal peptides (SP) (circles, black), lipid-binding peptides (LBP) (circles, blue), amphitropics (circles, green) and others (circles, orange) are depicted, see Table 2 for details.
 ), Globular (
), Globular (
 ) and TransMembrane (■) segments are depicted, see Table 1 for details. Examples of signal peptides (SP) (circles, black), lipid-binding peptides (LBP) (circles, blue), amphitropics (circles, green) and others (circles, orange) are depicted, see Table 2 for details.
) and TransMembrane (■) segments are depicted, see Table 1 for details. Examples of signal peptides (SP) (circles, black), lipid-binding peptides (LBP) (circles, blue), amphitropics (circles, green) and others (circles, orange) are depicted, see Table 2 for details.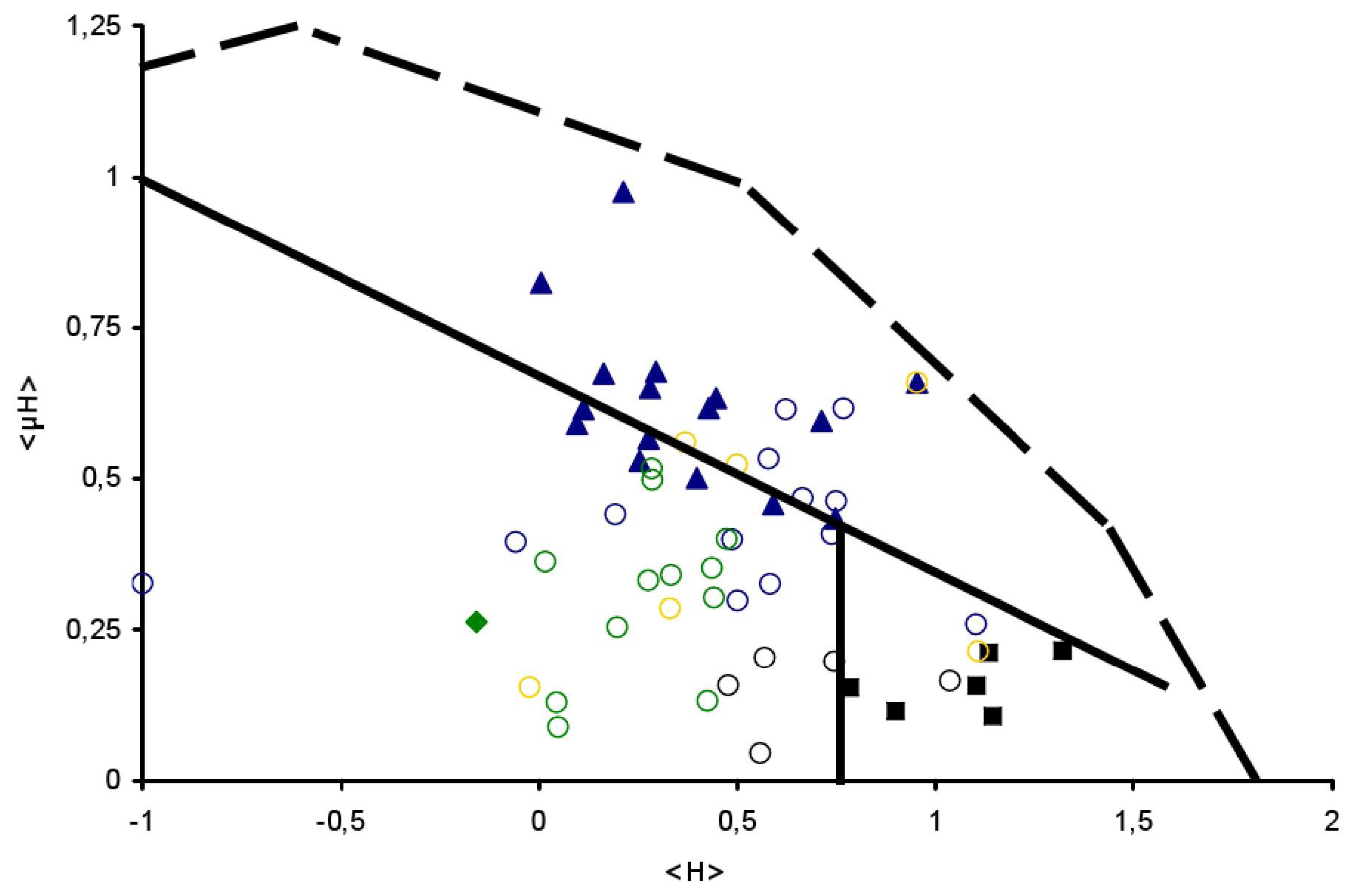
| Name | Sequence | z | <H> | <μH> | D | Conf. |
|---|---|---|---|---|---|---|
| RW16 | RRWRRWWRRWWRRWRR | 10 | 0.213 | 0.975 | YES | S [17] |
| RL16 | RRLRRLLRRLLRRLRR | 10 | 0.006 | 0.824 | YES | S [17] |
| Pbuy | FRKLFRVYSNFLRGKLKL | 6 | 0.280 | 0.650 | YES | S [18] |
| Pill | KQLIRFLKRLDRNLWGLA | 4 | 0.447 | 0.633 | YES | S [18] |
| Pc9k | NRLARHFRDIAGRVNQRL | 4 | 0.096 | 0.591 | YES | S [18] |
| Pqc7 | LKDVEEAQQKIINIIRRL | 1 | 0.280 | 0.650 | YES | S [18] |
| Pc3c | WYSEMKRNVQRLERAIEE | 0 | 0.113 | 0.615 | NO | S [18] |
| Pihf | RDAKELVELFFEEIRRAL | −1 | 0.276 | 0.566 | NO | S [18] |
| KL | KLLKLLLKLLKLLLKLLL | 5 | 0.953 | 0.659 | YES | S [19] |
| CRAMP18 | GEKLKKIGQKIKNFFQKL | 5 | 0.164 | 0.674 | YES | S [19] |
| SPLN14–27 | SLSRYAKLANRLA | 3 | 0.254 | 0.530 | YES | S [20] |
| SPLN28–41 | PKLLETFLSKWIG | 1 | 0.712 | 0.596 | YES | S [20] |
| Histatin 5 | SHAKRHHGYKRKFHEKHH | 5 | −0.157 | 0.263 | YES | G [21] |
| PGLaa | SKAGAIAGKIAKVALKAL | 3 | 0.398 | 0.501 | YES | S [21] |
| SP-B(7–24) | YCWLCRALIKRIQAMIPK | 4 | 0.747 | 0.434 | YES | S [22] |
| PC-TP196- | VPNFLKDMARACQNYLKK | 3 | 0.295 | 0.677 | YES | S [23] |
| Equinatoxin II | ASLSFDILKTVLEALGNV | −1 | 0.591 | 0.458 | NO | S [24] |
| KL4 | KLLLLKLLLLKLLLLKLL | 4 | 1.102 | 0.157 | YES | M [25] |
| KALP23 | KKLALALALALALALALA | 2 | 0.783 | 0.154 | YES | M [26] |
| WALP23 | WWLALALALALALALALA | 0 | 1.143 | 0.107 | NO | M [26] |
| Glycophorin A (92–114) | ITLIIFGVMAGVIGTILLI | 0 | 1.133 | 0.213 | NO | M [27] |
| TMX31 | WISFAISCFLLCVVLLGF | 0 | 1.321 | 0.216 | NO | M [28] |
| MHCClassII | VLVALLLAGQATTAYFLY | 0 | 0.899 | 0.115 | NO | M [29] |
| Name | Sequence | z | <H> | <μH> | D | Conf. |
|---|---|---|---|---|---|---|
| SP & LBP: | ||||||
| 1. prePhoE | KKSTLALVVMGIVASASV | 2 | 0.558 | 0.045 | Y | [31] |
| 2. preLamB | RKLPLAVAVAAGVMSAQA | 2 | 0.478 | 0.157 | Y | [32] |
| 3. proOmpA | KKTAIAIAVALAGFATVA | 2 | 0.569 | 0.204 | Y | [33] |
| 4. prePhoA | TIALALLPLLPTPVTKAR | 2 | 0.744 | 0.197 | Y | [34] |
| 5. Ovalbumin | IFYCPIAIMSALAMVTLG | 0 | 1.036 | 0.165 | N | [35] |
| 6. Aurein 1.2 | GLFDIKKVASVIGGL | 1 | 0.583 | 0.326 | N | [36,37] |
| 7. Citropin | GLFDVIKKVASVIGGL | 1 | 0.623 | 0.614 | Y | [36,37] |
| 8. Maculatin 1.1 | GLFGVLAKVAAHVVPAIA | 1 | 0.738 | 0.408 | Y | [36,37] |
| 9. VP1 | GTAMRILGGVI | 1 | 0.665 | 0.468 | Y | [38] |
| 10. HA2 FP | FGAIAGFIENGWEGMIDG | −3 | 0.579 | 0.533 | N | [38] |
| 11. AP1 | GEQGALAQFGEWL | −2 | 0.488 | 0.399 | N | [39] |
| 12. SIV peptide | GVFVLGFLGFLA | 0 | 1.102 | 0.259 | N | [40] |
| 13. Gaegurin 5 | LGALFKVASKVLPSVCAI | 2 | 0.749 | 0.463 | Y | [41] |
| 14. PBP5 | GNFFGKIIDYIKLMFHHW | 1 | 0.768 | 0.616 | Y | [42] |
| 16. Penetratin | RQIKIWFQNRRMKWKK | 7 | 0.193 | 0.327 | Y | [43] |
| 17. Polyarginine-R9 | RRRRRRRRR | 9 | −1.010 | 0.146 | Y | [44] |
| 18. Substance-P | RPKPQQFFGLM | 2 | 0.501 | 0.298 | Y | [45] |
| 19. Dermaseptin B2 | IKEVGKEAAKAAAKAAGK | 3 | −0.058 | 0.395 | Y | [46] |
| Amphitropics: | ||||||
| 20. SecA(1–21) | MLIKLLTKVFGSRNDRTL | 3 | 0.442 | 0.303 | Y | [14] |
| 21. SecA(108–125) | KTLTATLPAYLNALTGKG | 2 | 0.437 | 0.352 | Y | [47] |
| 22. SecA(593–614) | ALMRIFASDRVSGMMRKL | 3 | 0.425 | 0.131 | Y | [48] |
| 23. SecA(865–882) | AAAAALAAQTGERKVGRN | 2 | 0.049 | 0.088 | Y | [14] |
| 24. FtsY(1–18) | MAKEKKRGFFSWLGFGQK | 4 | 0.277 | 0.332 | Y | [49] |
| 25. FtsY(188–208) | KPTKEGFFARLKRSLLKT | 5 | 0.198 | 0.254 | Y | [50] |
| 26. Apocyt c2–21 | VEKGKKIFVQKCAQCHTV | 3 | 0.333 | 0.341 | Y | [51] |
| 27. Apocyt c80–101 | AGIKKKTEREDLIAYLKK | 3 | 0.046 | 0.129 | Y | [51] |
| 28. EcMinD251–269 | RPFRFIEEEKKGFLKRLF | 3 | 0.287 | 0.498 | Y | [52] |
| 29. α-synuclein1–15 | MDVFMKGLSKAKEGV | 1 | 0.285 | 0.517 | Y | [53] |
| 30. ARF1 | MGNIFANLFKGLFGKKEM | 2 | 0.474 | 0.400 | Y | [54] |
| 31. K-segment dehydrins | EKKGIMDKIKEKLPG | 2 | 0.017 | 0.363 | Y | [55] |
| Miscellaneous: | ||||||
| 32. Kes 1p (7–29) | SSSWTSFLKSIASFNGDL | 0 | 0.500 | 0.523 | N | [56] |
| 33. PBP4 | RRIPLVRFESRLYKDIYQNN | 3 | 0.331 | 0.285 | Y | [42] |
| 34. KCNQ1354–372 | KVQQKQRQKHFNRQIPAA | 5 | −0.023 | 0.154 | Y | [57] |
| 35. ABP280(49–71) | FTRWCNEHLKCVSKRIAN | 3 | 0.370 | 0.560 | Y | [58] |
| 36. L15K7 | KLLKLLLKLLKLLLKLLLKLLK | 5 | 0.953 | 0.659 | Y | [59] |
| Name | Sequence | z | <H> | <μH> | D | Confirmed |
|---|---|---|---|---|---|---|
| M13 coat protein: | ||||||
| 2KKSLVLKASVAVATLVPM19 | 3 | 0.559 | 0.072 | YES | [65] | |
| 47YAWAMVVVIVGATIGIKL64 | 1 | 0.923 | 0.062 | NO | [65] | |
| 54VIVGATIGIKLFKKFTSK71 | 4 | 0.553 | 0.288 | YES | - | |
| Ffh: | ||||||
| (P0AGD7) | 1MFDNLTDRLSRTLRNISG18 | 1 | 0.255 | 0.663 | YES | - |
| 44ALPVVREFINRVKEKAVG61 | 2 | 0.313 | 0.365 | YES | - | |
| 166QKPVDIVNAALKEAKLKF183 | 2 | 0.272 | 0.331 | YES | - | |
| 309SKVDRAQAEKLASKLKKG326 | 4 | −0.118 | 0.297 | YES | - | |
| 336EQLRQMKNMGGMASLMGK353 | 2 | 0.218 | 0.261 | YES | - | |
| 395KGSRKRRIAAGCGMQVQD412 | 4 | 0.008 | 0.140 | YES | - | |
| 415RLLKQFDDMQRMMKKMKK432 | 5 | 0.064 | 0.606 | YES | - | |
| 428KKMKKGGMAKMMRSMKGM445 | 7 | 0.039 | 0.327 | YES | - | |
| Fis1: | ||||||
| (P40515) | 35PTATIQSRFNYAWGLIKS52 | 2 | 0.514 | 0.349 | YES | [67] |
| 60LGVKILTDIYKEAESRRR77 | 2 | 0.147 | 0.326 | YES | - | |
| 108RNNKQVGALKSMVEDKIQ125 | 2 | 0.023 | 0.305 | YES | - | |
| 133VVAGGVLAGAVAVASFFL150 | 0 | 0.811 | 0.145 | YES | [67] |
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Keller, R.C.A. New User-Friendly Approach to Obtain an Eisenberg Plot and Its Use as a Practical Tool in Protein Sequence Analysis. Int. J. Mol. Sci. 2011, 12, 5577-5591. https://doi.org/10.3390/ijms12095577
Keller RCA. New User-Friendly Approach to Obtain an Eisenberg Plot and Its Use as a Practical Tool in Protein Sequence Analysis. International Journal of Molecular Sciences. 2011; 12(9):5577-5591. https://doi.org/10.3390/ijms12095577
Chicago/Turabian StyleKeller, Rob C.A. 2011. "New User-Friendly Approach to Obtain an Eisenberg Plot and Its Use as a Practical Tool in Protein Sequence Analysis" International Journal of Molecular Sciences 12, no. 9: 5577-5591. https://doi.org/10.3390/ijms12095577




