Estimation of the Genetic Diversity in Tetraploid Alfalfa Populations Based on RAPD Markers for Breeding Purposes
Abstract
:1. Introduction
2. Experimental Section
2.1. Plant Material and DNA Isolation
2.2. RAPD Analysis
2.3. Data Analysis
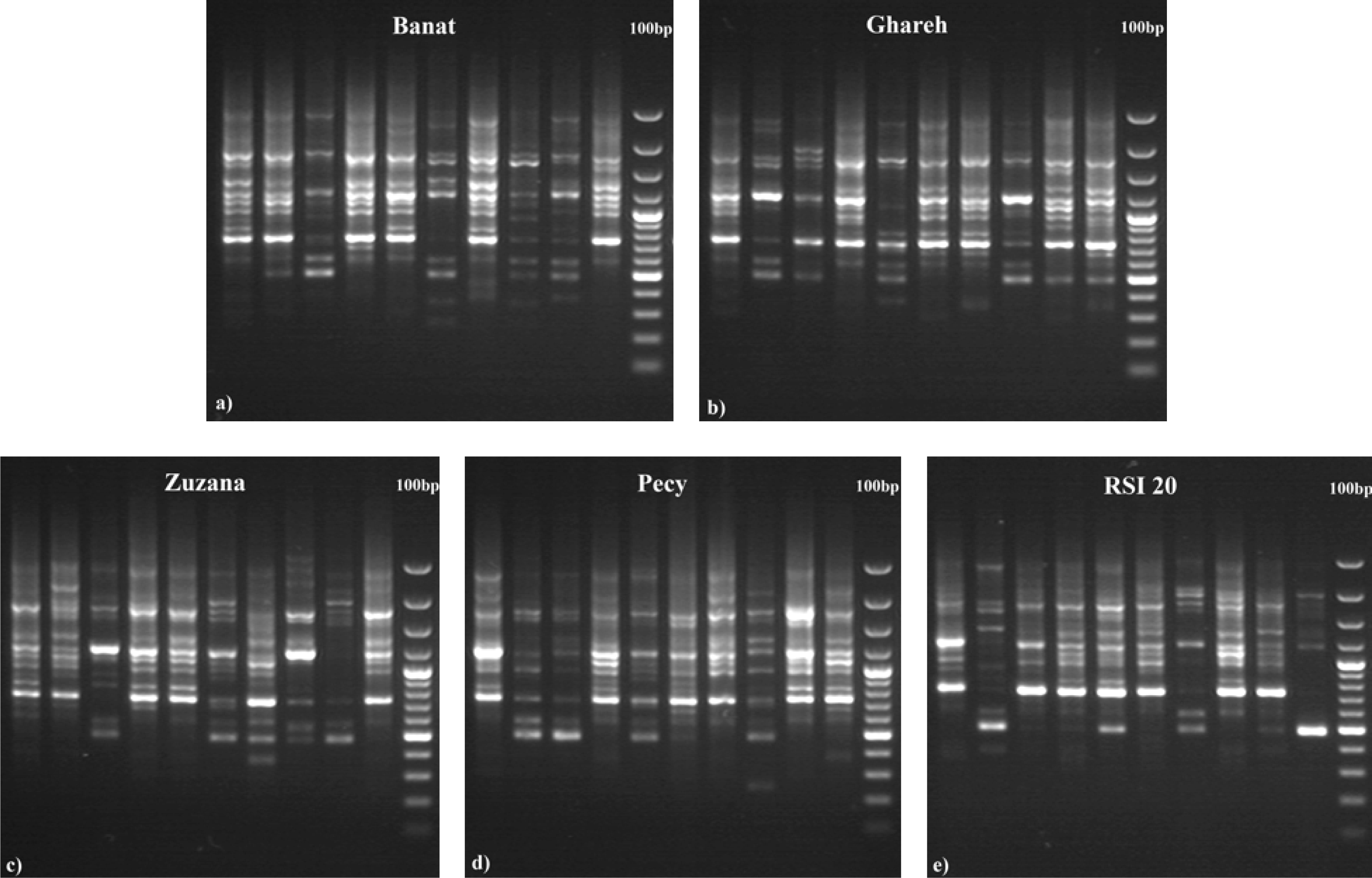
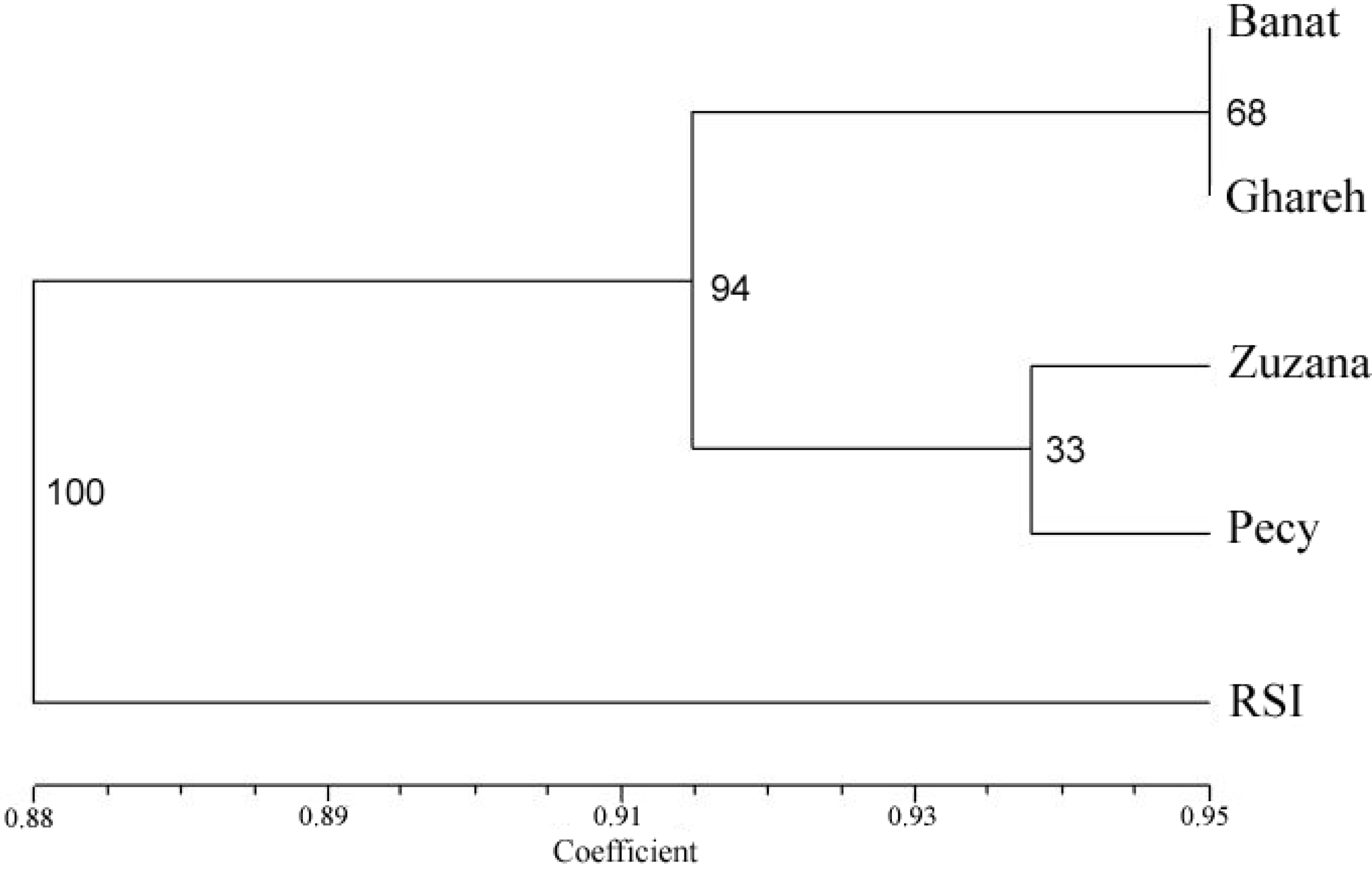
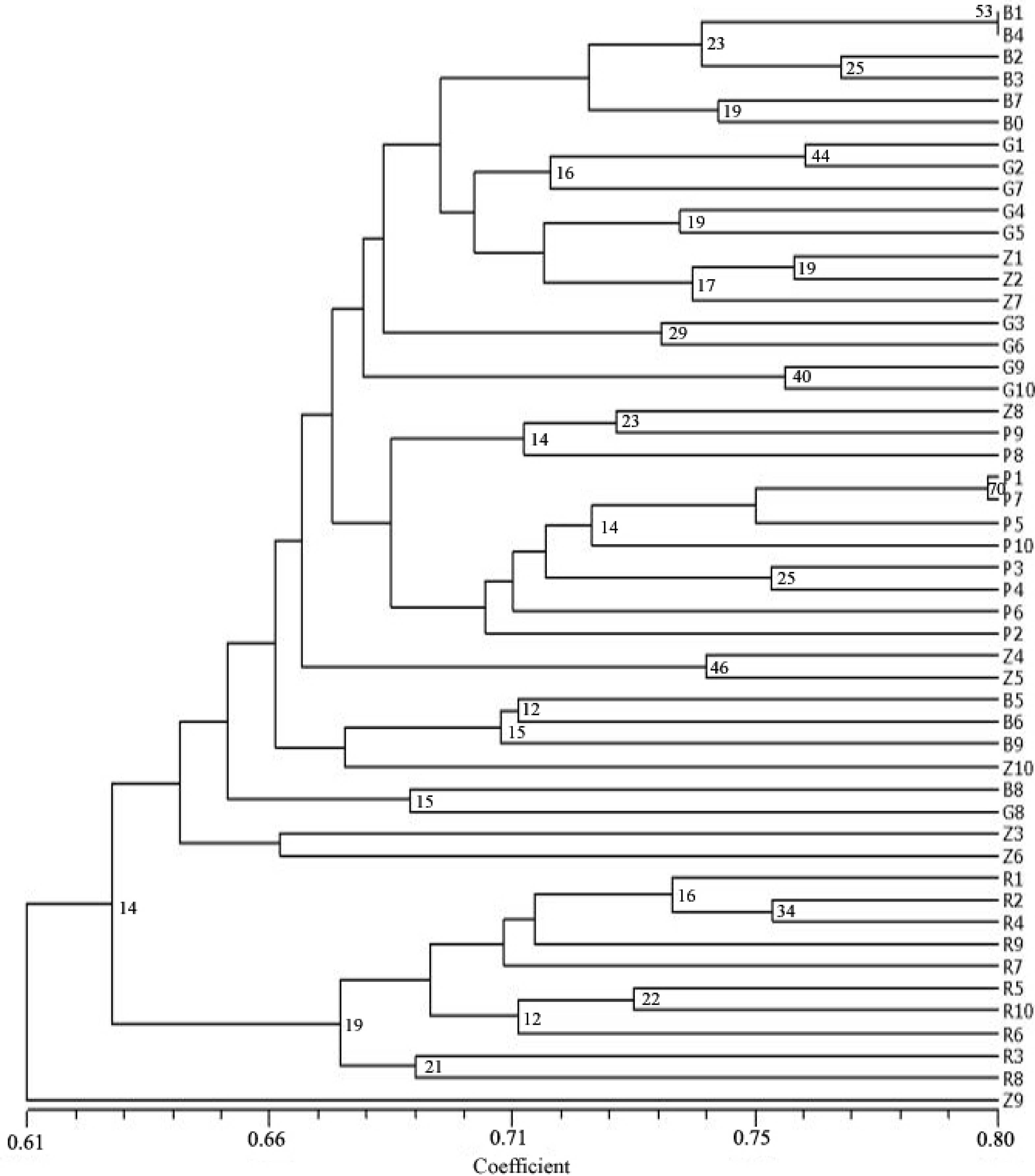
3. Results
4. Discussion
5. Conclusions
Acknowledgments
- Conflict of InterestThe authors declare no conflict of interest.
References
- Veronesi, F; Brummer, EC; Huyghe, C. Alfalfa. In Fodder Crops and Amenity Grasses. Series: Handbook of Plant Breeding; Boller, B, Posselt, UK, Veronesi, F, Eds.; Springer: New York, NY, USA, 2010; Volume 5, pp. 395–437. [Google Scholar]
- Julier, B; Barre, P; Hebert, Y; Huguet, T; Huyghe, C. Methodology of alfalfa breeding: A review of recent achievements. Czech J. Genet. Plant Breed 2003, 39, 75–77. [Google Scholar]
- Brummer, E. Applying genomics to alfalfa breeding programs. Crop Sci 2004, 44, 1904–1906. [Google Scholar]
- Rowe, DE; Hill, RR. Breeding theory and the development of alfalfa. The Alfalfa Genome, 1999. Available online: www.naaic.org/TAG/TAGpapers/RoweAbs.html (accessed on 17 August 2011).
- Julier, B; Flajoulot, S; Barre, P; Cardinet, G; Santoni, S; Huguet, T; Huyghe, C. Construction of two genetic linkage maps in cultivated tetraploid alfalfa (Medicago sativa) using microsatellite and AFLP markers. BMC Plant Biol 2003b, 3, 9. [Google Scholar]
- Milić, D; Katić, S; Karagić, Đ; Gvozdanović-Varga, J; Petrović, S; Boćanski, J. Genetic control of agronomic traits in alfalfa (M. sativa ssp. sativa L.). Euphytica 2011. [Google Scholar] [CrossRef]
- Tucak, M; Popovic, S; Cupic, T; Grljusic, S; Bolaric, S; Kozumplik, V. Genetic diversity of alfalfa (Medicago spp.) estimated by molecular markers and morphological characters. Period. Biol 2008, 110, 243–249. [Google Scholar]
- Gherardi, M; Mangin, B; Goffinet, B; Bonet, D; Huguet, T. A method to measure genetic distance between alogamous populations of alfalfa (Medicago sativa) using RAPD molecular markers. Theor. Appl. Genet 1998, 96, 406–412. [Google Scholar]
- Flajoulot, S; Ronfort, J; Baudouin, P; Barre, P; Huguet, T; Huyghe, C; Julier, B. Genetic diversity among alfalfa (Medicago sativa) cultivars coming from a breeding program, using SSR markers. Theor. Appl. Genet 2005, 111, 1420–1429. [Google Scholar]
- Vandemark, JG; Ariss, JJ; Bauchan, GA; Larsen, CR; Hughes, JT. Estimating genetic relationships among historical sourcesof alfalfa germplasm and selected cultivars with sequence related amplified polymorphisms. Euphytica 2006, 152, 9–16. [Google Scholar]
- Noeparvar, S; Valizadeh, M; Monirifar, H; Haghighi, A; Darbani, B. Genetic diversity among and within alfalfa populations native to Azarbaijan based on RAPD analysis. J. Biol. Res.-Thessalon 2008, 10, 159–169. [Google Scholar]
- Carelli, M; Gnocchi, G; Scotti, C. Alfalfa germplasm from Sahara oasis: Characterization by means of bio-agronomic traits and SSR markers. Plant Breed 2009, 128, 271–277. [Google Scholar]
- Morales, C; Martinez, C. Variation of PGM and IDH isozymes for identification of alfalfa varieties. Euphytica 2000, 112, 137–143. [Google Scholar]
- Pupilli, F; Labombarda, P; Scotti, C; Arcioni, S. RFLP analysis allows for the identification of alfalfa ecotypes. Plant Breed 2000, 119, 271–276. [Google Scholar]
- Falahati-Anbaran, M; Habashi, A; Esfahany, M; Mohammadi, S; Ghareyazie, B. Population genetic structure based on SSR markers in alfalfa (Medicago sativa L.) from various regions contiguous to the centers of origin of the species. J. Genet 2007, 86, 59–63. [Google Scholar]
- Yu, P; Pauls, KP. Segregation of random amplified polymorphic DNA markers and strategies for molecular mapping in tetraploid alfalfa. Genome 1993, 36, 844–851. [Google Scholar]
- Sledge, M; Ray, I; Jiang, G. An expressed sequence tag SSR map of tetraploid alfalfa (Medicago sativa L.). Theor. Appl. Genet 2005, 111, 980–992. [Google Scholar]
- Williams, JGK; Kubelik, AR; Livak, KJ; Rafalski, JA; Tingey, SV. DNA polymorphism amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res 1990, 18, 6531–6535. [Google Scholar]
- Melchinger, AE. Use of RFLP Markers for Analysis of Genetic Relationships among Breeding Materials and Prediction of Hybrid Performance. In International Crop Science; Buxton, DR, Shibles, R, Forsberg, RA, Blad, BL, Asay, KH, Paulson, GM, Wilson, RF, Eds.; Crop Science Society of America (CSSA): Medison, WI, USA, 1993; pp. 621–628. [Google Scholar]
- Milic, D; Katic, S; Gvozdanovic-Varga, J; Mikic, A; Karagic, DJ; Vasiljevic, S. Divergence of lucerne (Medicago sativa L.) genotypes for yield components. J. Sci. Agric. Res 2007, 68, 97–102. [Google Scholar]
- Somma, M. Extraction and Purification of DNA. In The Analysis of Food Samples for the Presence of Genetically Modified Organisms; special publication 1.03.114 ed.; Querci, M, Jermini, M, van den Eade, G, Eds.; European Commision, Joint Research Centre: Ispra, Italy, 2004; Chapter 4. [Google Scholar]
- Nagl, N; Taski-Ajdukovic, K; Barac, G; Milic, D; Katic, S. Research on possibility of RAPD markers application in detection of DNA polymorphism of alfalfa varieties. Field Veg. Crop Res 2010, 47, 511–516. [Google Scholar]
- Smith, J; Chin, E; Shu, H; Smith, O; Wall, S; Senior, M; Mitchell, S; Kresovich, S; Ziegle, J. An evaluation of the utility of SSR loci as a molecular markers in maize (Zea mays L.): Comparisons with data from RFLPs and pedigree. Theor. Appl. Genet 1997, 95, 163–173. [Google Scholar]
- Yeh, FC; Yang, RC; Boyle, T. POPGENE The User Friendly Software for Population Genetic Analysis; Molecular Biology and Biotechnology Center, University of Alberta: Edmonton, Canada, 1997. [Google Scholar]
- Kimura, M; Crow, J. The number of alleles that can be maintained in a finite population. Genetics 1964, 49, 725–738. [Google Scholar]
- Nei, M. Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. USA 1973, 70, 3321–3323. [Google Scholar]
- Lewontin, RC. The apportionment of human diversity. Evol. Biol 1972, 6, 381–398. [Google Scholar]
- Excoffier, L; Smouse, P; Quattro, G. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics 1992, 131, 479–491. [Google Scholar]
- Excoffier, L; Laval, G; Schneider, S. Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evol. Bioinf. Online 2005, 1, 47–50. [Google Scholar]
- Rohlf, FJ. NTSYSpc: Numerical Taxonomy and Multivariate Analysis System, Version 2.1a; Exeter Software: Setauket, NY, USA, 2000; p. 44. [Google Scholar]
- Mantel, NA. The detection of disease clustering and a generalized regression approach. Cancer Res 1967, 27, 209–220. [Google Scholar]
- Pavlicek, A; Hrda, S; Flegr, J. Free-Tree—freeware program for construction of phylogenic trees on the basis of distance data and bootstrap/jackknife analysis of the tree robustness. Application in the RAPD analysis of the genus Frankelia. Folia Biol 1999, 45, 97–99. [Google Scholar]
- STATSOFT. Statistica: Data analysis software system. Version 8. Available online: http://www.statsoft.com/ (accessed on 5 August 2010).
- Mengoni, A; Gori, A; Bazzicalupo, M. Use of RAPD and microsatellite (SSR) variation to assess genetic relationships among populations of tetraploid alfalfa Medicago sativa. Plant Breed 2000, 119, 311–317. [Google Scholar]
- Crochemore, M; Huyghe, C; Kerlan, M; Durand, F; Julier, B. Partitioning and distribution of RAPD variation in a set of populations of the Medicago sativa complex. Agronomie 1997, 16, 421–432. [Google Scholar]
- Kidwell, K; Bingham, E; Woodfield, D; Osborn, T. Molecular marker diversity and yield of isogenic 2× and 4× single-crosses of alfalfa. Crop Sci 1994, 34, 784–788. [Google Scholar]
- Kidwell, K; Hartweck, M; Yandell, S; Crump, M; Brummer, E; Moutray, E; Osborn, C. Forage yields of alfalfa populations derived from parents selected on the basis of molecular markers diversity. Crop Sci 1999, 39, 223–227. [Google Scholar]
- Riday, H; Brummer, E; Campbell, A; Luth, D; Cazcarro, P. Comparisons of genetic and morphological distance with heterosis between Medicago sativa subsp. sativa and subsp. falcata. Theor. Appl. Genet 2003, 131, 37–45. [Google Scholar]
- Maureira, I; Ortega, F; Campos, H; Osborn, C. Population structure and combining ability of diverse Medicago sativa germplasms. Theor. Appl. Genet 2004, 109, 775–782. [Google Scholar]
| Variety | Description |
|---|---|
| Banat (NS Banat ZMS II) | An old variety developed at the Institute of Field and Vegetable Crops, Novi Sad, Serbia by individual selection from local populations (Pannonian type of alfalfa). Has rapid initial growth and fast regrowth after cutting. Plant height at early flowering is over 80 cm. Proportion of leaves in green forage yield is 450–500 g kg−1. Green forage yield is about 80 t ha−1, hay yield is 15–20 t ha−1. Resistant to drought, low temperatures and frequent cuttings. |
| Ghareh (Ghareh Yon Geh) | Variety developed at the Institute Karaj in Iran, the center of alfalfa origin. Well adapted to agroecological conditions of Serbia (resistant to drought and low temperature). Has tall, large plants that regrow fast after cutting (37.1 cm) and high hay yield (16–19 t ha−1). |
| Zuzana | An old variety developed at the breeding station in Zelezice, Brno, Czech Republic, with good adaptation to agroecological conditions of Serbia. Has good dry matter yield (14 t ha−1), a larger number of shorter internodes (tolerance to lodging), slower regrowth after cutting (higher dormancy), and higher susceptibility to drought. Represents a transition between Pannonian and Western European type of varieties. |
| Pecy | An old French variety developed by company R2N. A typical variety of the Western European type. Has good resistance to lodging and main alfalfa diseases. Well able to withstand low temperatures but is susceptible to drought. Has an exceptional quality, with high proportion of leaves in yield (48–56%), and larger number (13) of short internodes (5.2 cm). |
| RSI 20 | Variety originating from Spain. Early-maturing variety with high dry matter yield (17.9 t ha−1) and excellent quality (crude protein content of 22.2%). Has low dormancy (fast regrowth after cutting −39.4 cm), tolerance to high temperatures and drought, but it is sensitive to cold. Because of smaller number (10) of long internodes (6.3 cm) it is susceptible to lodging. |
| Primer | Sequence (5′-3′) | Max. No. of Bands | Band Size Range (bp) | PIC |
|---|---|---|---|---|
| X07 | GAGCGAGGCT | 14 | 400–2000 | 0.267 |
| X09 | GGTCTGGTTG | 12 | 600–3000 | 0.286 |
| X12 | TCGCCAGCCA | 13 | 400–2000 | 0.174 |
| X17 | GACACGGACC | 17 | 400–3000 | 0.368 |
| Y02 | CATCGCCGCA | 7 | 900–3500 | 0.314 |
| Y05 | GGCTGCGACA | 11 | 700–4000 | 0.285 |
| Y06 | AAGGCTCACC | 8 | 1000–3500 | 0.358 |
| Y07 | AGAGCCGTCA | 14 | 700–4000 | 0.291 |
| Y10 | CAAACGTGGG | 9 | 650–4000 | 0.320 |
| Y11 | AGACGATGGG | 13 | 300–2500 | 0.308 |
| Y13 | GGGTCTCGGT | 10 | 500–2800 | 0.345 |
| Y15 | AGTCGCCCTT | 9 | 350–3000 | 0.361 |
| Z01 | TCTGTGCCAA | 6 | 450–1600 | 0.011 |
| Z07 | CCAGGAGGAC | 11 | 600–3000 | 0.158 |
| Z12 | TCAACGGGAC | 8 | 1000–6000 | 0.242 |
| Z14 | TCGGAGGTTC | 8 | 900–3500 | 0.256 |
| Z17 | CCTTCCCACT | 10 | 400–5000 | 0.374 |
| Variety | P (No.) | P (%) | Ne | He | I |
|---|---|---|---|---|---|
| Banat | 90 | 57.69 | 1.389 ± 0.400 | 0.220 ± 0.212 | 0.323 ± 0.300 |
| Ghareh | 88 | 56.41 | 1.387 ± 0.389 | 0.217 ± 0.209 | 0.319 ± 0.298 |
| Zuzana | 104 | 66.67 | 1.453 ± 0.399 | 0.256 ± 0.208 | 0.375 ± 0.292 |
| Pecy | 88 | 56.41 | 1.393 ± 0.407 | 0.220 ± 0.215 | 0.322 ± 0.305 |
| RSI 20 | 87 | 55.77 | 1.385 ± 0.402 | 0.217 ± 0.213 | 0.318 ± 0.302 |
| Mean | 91.4 | 58.59 | 1.399 ± 0.399 | 0.226 ± 0.211 | 0.322 ± 0.299 |
| Overall | 129 | 82.69 | 1.498 ± 0.377 | 0.286 ± 0.189 | 0.426 ± 0.257 |
| Source of Variation | Df | SSD | Variance Components | Percentage Variation | P |
|---|---|---|---|---|---|
| Among populations | 4 | 143.464 | 2.03623 | 11.61 | <10−5 |
| Within populations | 45 | 697.666 | 15.5369 | 88.39 | <10−5 |
| Total | 841.130 | 17.53992 | |||
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Nagl, N.; Taski-Ajdukovic, K.; Barac, G.; Baburski, A.; Seccareccia, I.; Milic, D.; Katic, S. Estimation of the Genetic Diversity in Tetraploid Alfalfa Populations Based on RAPD Markers for Breeding Purposes. Int. J. Mol. Sci. 2011, 12, 5449-5460. https://doi.org/10.3390/ijms12085449
Nagl N, Taski-Ajdukovic K, Barac G, Baburski A, Seccareccia I, Milic D, Katic S. Estimation of the Genetic Diversity in Tetraploid Alfalfa Populations Based on RAPD Markers for Breeding Purposes. International Journal of Molecular Sciences. 2011; 12(8):5449-5460. https://doi.org/10.3390/ijms12085449
Chicago/Turabian StyleNagl, Nevena, Ksenija Taski-Ajdukovic, Goran Barac, Aleksandar Baburski, Ivana Seccareccia, Dragan Milic, and Slobodan Katic. 2011. "Estimation of the Genetic Diversity in Tetraploid Alfalfa Populations Based on RAPD Markers for Breeding Purposes" International Journal of Molecular Sciences 12, no. 8: 5449-5460. https://doi.org/10.3390/ijms12085449




