Investigations on Inhibitors of Hedgehog Signal Pathway: A Quantitative Structure-Activity Relationship Study
Abstract
:1. Introduction
2. Results and Discussion
2.1. The Influence of Descriptors on the QSAR Modeling of Inhibitors of Hedgehog Signal Pathway
2.2. The Influence of Data Division on the QSAR Modeling of Inhibitors of Hedgehog Signal Pathway
2.3. Comparison of PLS and SVR for QSAR Data Regression
2.4. Comparison of Binary Bayesian Inference and SVM for QSAR Data Classification
2.5. Cell Line Analysis
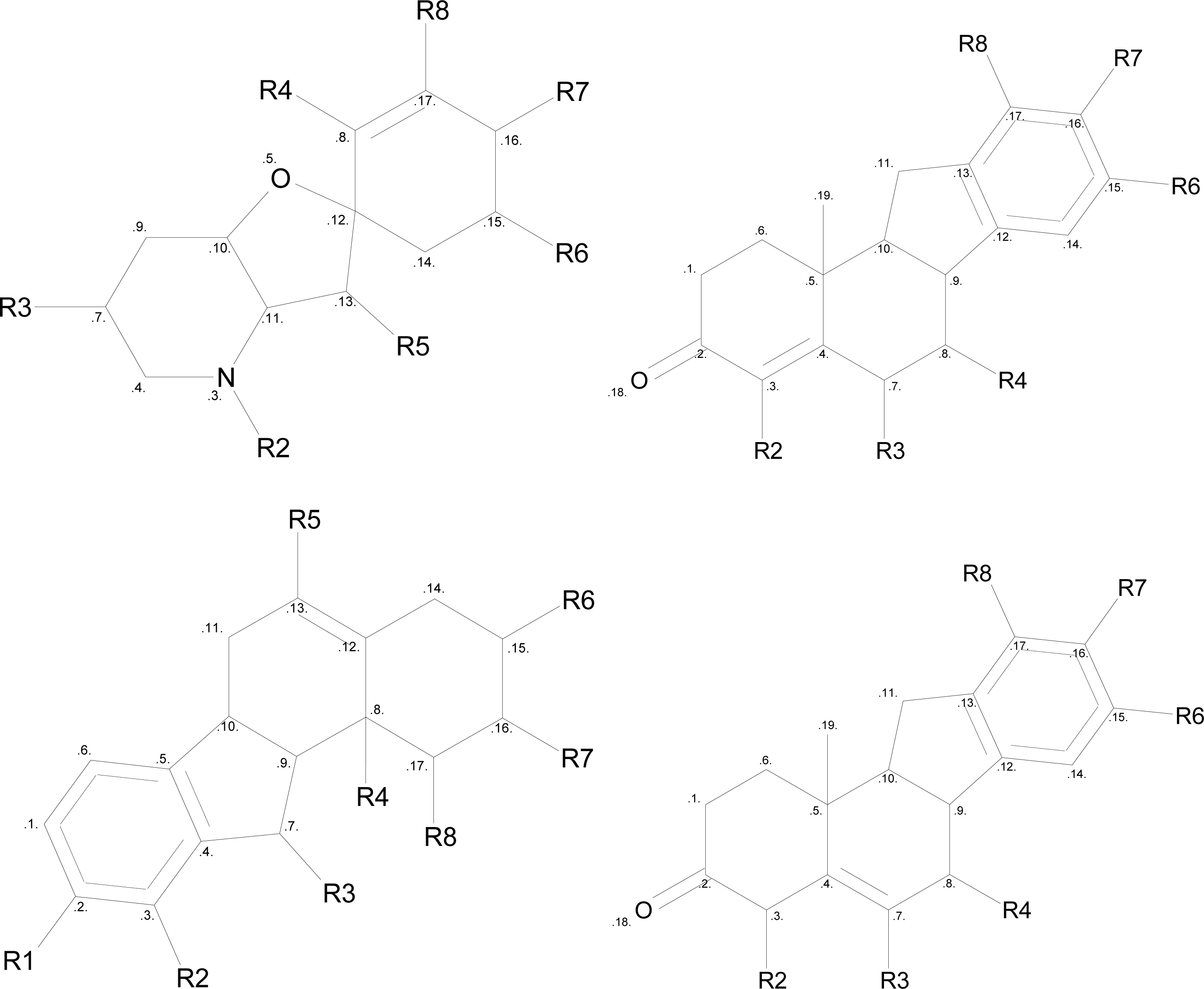
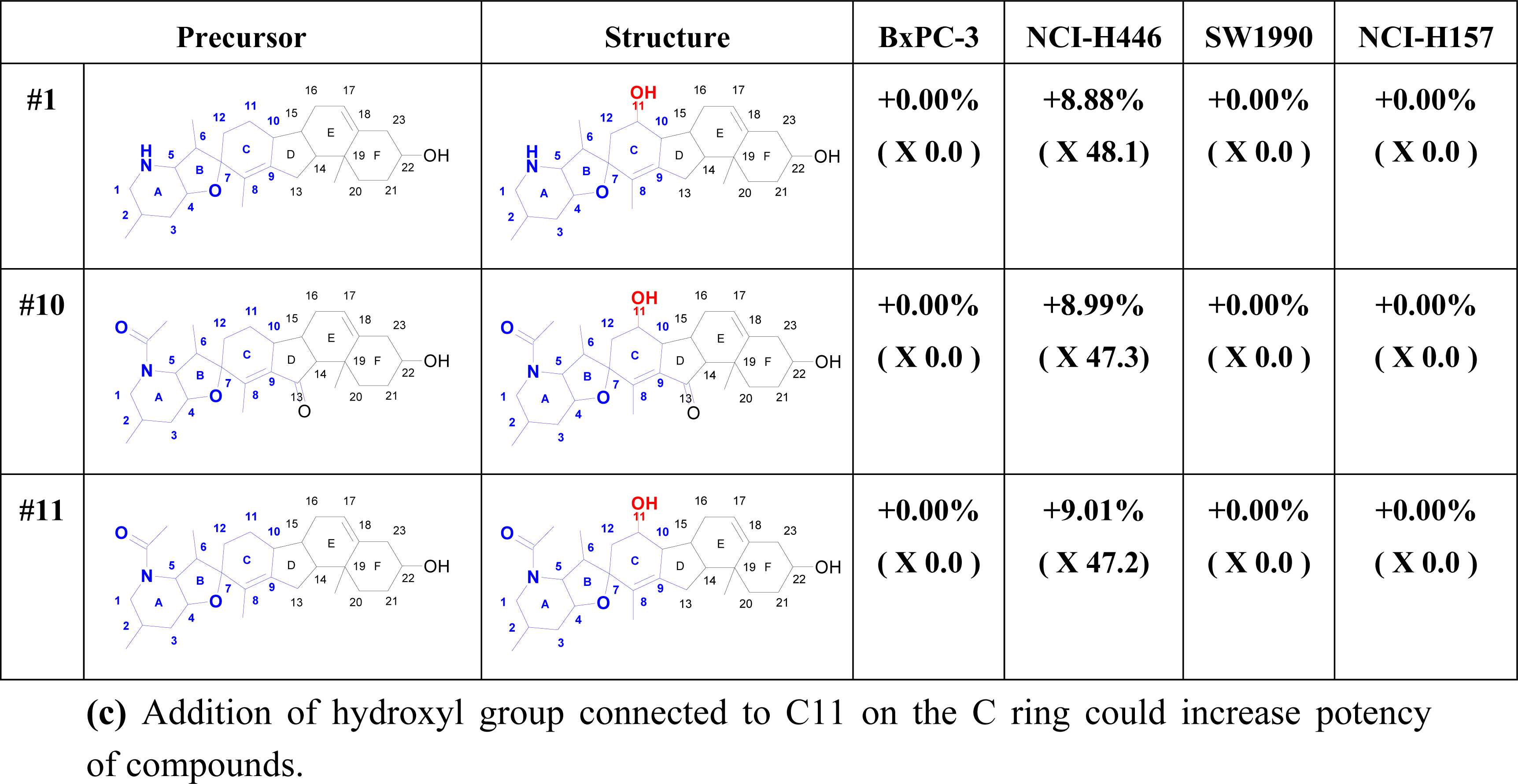
2.6. Structure Activity Report
3. Material and Methods
3.1. Dataset and Data Division Methods
3.1.1. Diverse Subset
3.1.2. Cluster plus Diverse Subset
3.2. Structural Descriptors
3.3. Statistic Modeling
3.3.1. PLS Method
3.3.2. SVR
3.3.3. Binary Bayesian Inference
- Estimates two distributions: one for the active compounds and one for the inactive ones in the training set. The separation of active and inactive sets is manually defined by a Binary Threshold.
- Counts the frequency of occurrence of a particular descriptor value in active and inactive cases.
- Accumulates a histogram of the observed sample values over the classes. The distribution is convoluted with a Gaussian (σ = 0.25, the smoothing width) to avoid sensitivity to bin boundaries.
- A histogram of property distributions is derived for each descriptor for “active ” and “inactive” (yes/no) sets. Those descriptors which differentiate the two sets will have a high impact in the model, those which do not, will drop out.
3.3.4. SVM Classification
3.4. SAReport
4. Conclusions
Supplementary Materials
ijms-12-03018-s001.docAbbreviations
| R2 = | correlation coefficient in self fitting of training data set |
| Q2 = | correlation coefficient in cross validation fitting of training data set |
| r2 = | correlation coefficient in fitting of test data set |
| A = | percentage accuracy of binary model = Total accuracy |
| A0 = | percentage accuracy of inactive subset |
| A1 = | percentage accuracy of active subset |
| At = | A in self fitting of training data set |
| Av = | A in cross validation fitting of training data set |
| Ap = | A in fitting of test data set |
| DLI = | Drug-like Index |
| PLS = | Partial Least Squares |
| SVR = | Support Vector Regression |
| SVM = | Support Vector Machine |
| ANN = | Artificial Neural Networks |
| SAReport = | Structure-Activity Report |
Acknowledgments
References
- Ingham, PW; McMahon, AP. Hedgehog signaling in animal development: Paradigms and principles. Gene. Dev 2001, 15, 3059–3087. [Google Scholar]
- Oro, AE; Higgins, KM; Hu, Z; Bonifas, JM; Epstein, EH, Jr; Scott, MP. Basal cell carcinomas in mice overexpressing sonic hedgehog. Science 1997, 276, 817–821. [Google Scholar]
- Kinzler, KW; Bigner, SH; Bigner, DD; Trent, JM; Law, ML; O’Brien, SJ; Wong, AJ; Vogelstein, B. Identification of an amplified, highly expressed gene in a human glioma. Science 1987, 236, 70–73. [Google Scholar]
- Dahmane, N; Lee, J; Robins, P; Heller, P; Ruiz i Altaba, A. Activation of the transcription factor Gli1 and the Sonic hedgehog signalling pathway in skin tumours. Nature 1997, 389, 876–881. [Google Scholar]
- Grachtchouk, M; Mo, R; Yu, S; Zhang, X; Sasaki, H; Hui, CC; Dlugosz, AA. Basal cell carcinomas in mice overexpressing Gli2 in skin. Nat. Genet 2000, 24, 216–217. [Google Scholar]
- Reifenberger, J; Wolter, M; Weber, RG; Megahed, M; Ruzicka, T; Lichter, P; Reifenberger, G. Missense mutations in SMOH in sporadic basal cell carcinomas of the skin and primitive neuroectodermal tumors of the central nervous system. Cancer Res 1998, 58, 1798–1803. [Google Scholar]
- Dahmane, N; Sanchez, P; Gitton, Y; Palma, V; Sun, T; Beyna, M; Weiner, H; Ruiz i Altaba, A. The Sonic Hedgehog-Gli pathway regulates dorsal brain growth and tumorigenesis. Development 2001, 128, 5201–5212. [Google Scholar]
- Chen, JK; Taipale, J; Young, KE; Maiti, T; Beachy, PA. Small molecule modulation of Smoothened activity. Proc. Nat. Acad. Sci. USA 2002, 99, 14071–14076. [Google Scholar]
- Chen, JK; Taipale, J; Cooper, MK; Beachy, PA. Inhibition of hedgehog signaling by direct binding of cyclopamine to smoothened. Gene. Dev 2002, 16, 2743–2748. [Google Scholar]
- Beachy, P; Porter, J. Hedgehog-derived polypeptides. US Patent No. 6911528, 28 June 2005.
- Taipale, J; Chen, JK; Cooper, MK; Wang, B; Mann, RK; Milenkovic, L; Scott, MP; Beachy, PA. Effects of oncogenic mutations in smoothened and patched can be reversed by cyclopamine. Nature 2000, 406, 1005–1009. [Google Scholar]
- Giannis, A; Heretsch, P; Sarli, V; Stossel, A. Synthesis of cyclopamine using a biomimetic and diastereoselective approach. Angew. Chem. Int. Ed. Engl 2009, 48, 7911–7914. [Google Scholar]
- Zhang, J; Garrossian, M; Gardner, D; Garrossian, A; Chang, YT; Kim, YK; Chang, CW. Synthesis and anticancer activity studies of cyclopamine derivatives. Bioorg. Med. Chem. Lett 2008, 18, 1359–1363. [Google Scholar]
- Janardanannair, S; Adams, J; Ripka, AS. Methods for preparation cyclopamine analogs and use thereof in treating cancers. U.S. Patent 7,407,967 B2, 5 August 2008.
- Tang, J; Li, HL; Shen, YH; Jin, HZ; Yan, SK; Liu, RH; Zhang, WD. Antitumor activity of extracts and compounds from the rhizomes of Veratrum dahuricum. Phytother. Res 2008, 22, 1093–1096. [Google Scholar]
- Tang, J; Li, HL; Shen, YH; Jin, HZ; Yan, SK; Liu, XH; Zeng, HW; Liu, RH; Tan, YX; Zhang, WD. Antitumor and antiplatelet activity of alkaloids from Veratrum dahuricum. Phytother. Res 2010, 24, 821–826. [Google Scholar]
- Xu, J; Stevenson, J. Drug-like index: A new approach to measure drug-like compounds and their diversity. J. Chem. Inf. Comput. Sci 2000, 40, 1177–1187. [Google Scholar]
- Labute, P. A widely applicable set of descriptors. J. Mol. Graph. Model 2000, 18, 464–477. [Google Scholar]
- Molecular Operation Eenvironment, version 200810; Chemical Computing Group Inc: Montreal, Quebec, Canada, 2008.
- Discovery Studio, version 20; Accelrys Software Inc.: San Diego, CA, USA, 2007.
- Sybyl, version 68; Tripos Inc: St Louis, MO, USA, 2001.
- Balabin, RM; Lomakina, EI. Neural network approach to quantum-chemistry data: Accurate prediction of density functional theory energies. J. Chem. Phys 2009, 131, 74104. [Google Scholar]
- Gelman, A; Carlin, JB; Stern, HS; Rubin, DB. Bayesian Data Analysis, 2nd ed; Chapman and Hall/CRC: Boca Raton, FL, USA, 2003. [Google Scholar]
- Ho, TK. A data complexity analysis of comparative advantages of decision forest constructors. Pattern Anal. Appl 2002, 5, 102–112. [Google Scholar]
- Cortes, C; Vapnik, V. Support-vector networks. Mach. Learn 1995, 20, 273–297. [Google Scholar]
- Patra, JC; Singh, O. Artificial neural networks-based approach to design ARIs using QSAR for diabetes mellitus. J. Comput. Chem 2009, 30, 2494–2508. [Google Scholar]
- Bucinski, A; Socha, A; Wnuk, M; Baczek, T; Nowaczyk, A; Krysinski, J; Gorynski, K; Koba, M. Artificial neural networks in prediction of antifungal activity of a series of pyridine derivatives against Candida albicans. J. Microbiol. Meth 2009, 76, 25–29. [Google Scholar]
- Kahn, I; Sild, S; Maran, U. Modeling the toxicity of chemicals to Tetrahymena pyriformis using heuristic multilinear regression and heuristic back-propagation neural networks. J. Chem. Inf. Model 2007, 47, 2271–2279. [Google Scholar]
- Vijayan, RS; Bera, I; Prabu, M; Saha, S; Ghoshal, N. Combinatorial library enumeration and lead hopping using comparative interaction fingerprint analysis and classical 2D QSAR methods for seeking novel GABA(A) alpha(3) modulators. J. Chem. Inf. Model 2009, 49, 2498–2511. [Google Scholar]
- Tang, H; Wang, XS; Huang, XP; Roth, BL; Butler, KV; Kozikowski, AP; Jung, M; Tropsha, A. Novel inhibitors of human histone deacetylase (HDAC) identified by QSAR modeling of known inhibitors, virtual screening, and experimental validation. J. Chem. Inf. Model 2009, 49, 461–476. [Google Scholar]
- Burden, FR; Winkler, DA. Optimal sparse descriptor selection for QSAR using bayesian methods. QSAR Comb. Sci 2009, 28, 645–653. [Google Scholar]
- Abdoa, A; Salima, N. Similarity-based virtual screening using bayesian inference network: Enhanced search using 2D fingerprints and multiple reference structures. QSAR Comb. Sci 2009, 28, 654–663. [Google Scholar]
- Li, Y; Wang, Y; Ding, J; Wang, Y; Chang, YQ; Zhang, SW. In silico prediction of androgenic and nonandrogenic compounds using random forest. QSAR Comb. Sci 2009, 28, 396–405. [Google Scholar]
- Zhu, JX; Lu, WC; Liu, L; Gu, TH; Niu, B. Classification of Src Kinase inhibitors based on support vector machine. QSAR Comb. Sci 2009, 28, 719–727. [Google Scholar]
- Polishchuk, PG; Muratov, EN; Artemenko, AG; Kolumbin, OG; Muratov, NN; Kuz’min, VE. Application of random forest approach to QSAR prediction of aquatic toxicity. J. Chem. Inf. Model 2009, 49, 2481–2488. [Google Scholar]
- Sun, M; Zheng, YG; Wei, HT; Chen, JQ; Cai, J; Ji, M. enhanced replacement method-based quantitative structure-activity relationship modeling and support vector machine classification of 4-Anilino-3-quinolinecarbonitriles as Src Kinase inhibitors. QSAR Comb. Sci 2009, 28, 312–324. [Google Scholar]
- Darnag, R; Schmitzer, A; Belmiloud, Y; Villemin, D; Jarid, A; Chait, A; Seyagh, M; Cherqaoui, D. QSAR studies of HEPT derivatives using support vector machines. QSAR Comb. Sci 2009, 28, 709–718. [Google Scholar]
- Rao, HB; Yang, GB; Tan, NX; Li, P; Li, ZR; Li, XY. Prediction of HIV-1 Protease inhibitors using machine learning approaches. QSAR Comb. Sci 2009, 28, 1346–1357. [Google Scholar]
- Goodarzi, M; Freitas, MP; Jensen, R. Feature selection and linear/nonlinear regression methods for the accurate prediction of glycogen synthase kinase-3beta inhibitory activities. J. Chem. Inf. Model 2009, 49, 824–832. [Google Scholar]
- Watkins, DN; Berman, DM; Burkholder, SG; Wang, B; Beachy, PA; Baylin, SB. Hedgehog signalling within airway epithelial progenitors and in small-cell lung cancer. Nature 2003, 422, 313–317. [Google Scholar]
- Thayer, SP; di Magliano, MP; Heiser, PW; Nielsen, CM; Roberts, DJ; Lauwers, GY; Qi, YP; Gysin, S; Fernandez-del Castillo, C; Yajnik, V; et al. Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature 2003, 425, 851–856. [Google Scholar]
- Sasai, K; Romer, JT; Lee, Y; Finkelstein, D; Fuller, C; McKinnon, PJ; Curran, T. Shh pathway activity is down-regulated in cultured medulloblastoma cells: Implications for preclinical studies. Cancer Res 2006, 66, 4215–4222. [Google Scholar]
- Helland, IS. On the structure of partial least squares regression. Comm. Stat. Simulat. Comput 1988, 17, 581–607. [Google Scholar]
- Gelaldi, P; Kowalski, R. Partial least squares regression: A tutorial. Anal. Chim. Acta 1986, 185, 1–17. [Google Scholar]
- Drucker, H; Burges, CJC; Kaufman, L; Smola, AJ; Vapnik, V. Support Vector Regression Machines; MIT Press: Cambridge, MA, USA, 1996; pp. 155–161. [Google Scholar]
- Watanabe, N; Adachi, H; Takase, Y; Ozaki, H; Matsukura, M; Miyazaki, K; Ishibashi, K; Ishihara, H; Kodama, K; Nishino, M; et al. 4-(3-Chloro-4-methoxybenzyl)aminophthalazines: Synthesis and inhibitory activity toward phosphodiesterase 5. J. Med. Chem 2000, 43, 2523–2529. [Google Scholar]
- Schuffenhauer, A; Ertl, P; Roggo, S; Wetzel, S; Koch, MA; Waldmann, H. The scaffold tree-visualization of the scaffold universe by hierarchical scaffold classification. J. Chem. Inf. Model 2007, 47, 47–58. [Google Scholar]



| BxPC-3 | NCI-H446 | SW1990 | NCI-H157 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| General | Drug-like | σ | General | Drug-like | σ | General | Drug-like | σ | General | Drug-like | σ | ||
| PLS | R2 | 0.552 | 0.494 | −0.058 | 0.659 | 0.526 | −0.133 | 0.644 | 0.585 | −0.059 | 0.527 | 0.531 | 0.004 |
| Q2 | 0.000 | 0.035 | 0.035 | 0.001 | 0.026 | 0.025 | 0.021 | 0.158 | 0.137 | 0.038 | 0.106 | 0.068 | |
| r2 | 0.102 | 0.307 | 0.205 | 0.218 | 0.025 | −0.193 | 0.084 | 0.193 | 0.109 | 0.019 | 0.118 | 0.099 | |
| SVR | R2 | 0.994 | 0.686 | 0.308 | 0.966 | 0.763 | −0.203 | 0.993 | 0.808 | −0.185 | 0.988 | 0.705 | −0.283 |
| Q2 | 0.994 | 0.000 | −0.994 | 0.962 | 0.002 | −0.96 | 0.992 | 0.069 | −0.923 | 0.987 | 0.001 | −0.986 | |
| r2 | 0.000 | 0.396 | 0.396 | 0.088 | 0.110 | 0.022 | 0.025 | 0.258 | 0.233 | 0.023 | 0.077 | 0.054 | |
| Bayesian inference | At | 0.883 | 0.917 | 0.034 | 1.000 | 0.967 | −0.033 | 0.900 | 0.933 | 0.033 | 0.967 | 0.933 | −0.034 |
| Av | 0.783 | 0.817 | 0.034 | 0.917 | 0.917 | 0 | 0.883 | 0.783 | −0.1 | 0.867 | 0.867 | 0 | |
| Ap | 0.606 | 0.576 | −0.03 | 0.758 | 0.879 | 0.121 | 0.576 | 0.667 | 0.091 | 0.485 | 0.636 | 0.151 | |
| SVM classification | At | 1.000 | 1.000 | 0 | 1.000 | 1.000 | 0 | 1.000 | 1.000 | 0 | 1.000 | 1.000 | 0 |
| Av | 0.550 | 0.500 | −0.05 | 0.867 | 0.817 | −0.05 | 0.650 | 0.533 | −0.117 | 0.633 | 0.617 | −0.016 | |
| Ap | 0.455 | 0.636 | 0.181 | 0.788 | 0.879 | 0.091 | 0.545 | 0.758 | 0.213 | 0.697 | 0.636 | −0.061 | |
| BxPC-3 | NCI-H446 | SW1990 | NCI-H157 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| General | Drug-like | σ | General | Drug-like | σ | General | Drug-like | σ | General | Drug-like | σ | ||
| PLS | R2 | 0.506 | 0.474 | −0.032 | 0.593 | 0.396 | −0.197 | 0.542 | 0.493 | −0.049 | 0.587 | 0.542 | −0.045 |
| Q2 | 0.011 | 0.007 | −0.004 | 0.015 | 0.019 | 0.004 | 0.005 | 0.002 | −0.003 | 0.006 | 0.040 | 0.034 | |
| r2 | 0.178 | 0.215 | 0.037 | 0.055 | 0.201 | 0.146 | 0.000 | 0.222 | 0.222 | 0.087 | 0.056 | −0.031 | |
| SVR | R2 | 0.997 | 0.716 | −0.281 | 0.965 | 0.756 | −0.209 | 0.993 | 0.839 | −0.154 | 0.987 | 0.655 | −0.332 |
| Q2 | 0.997 | 0.021 | −0.976 | 0.962 | 0.025 | −0.937 | 0.993 | 0.124 | −0.869 | 0.986 | 0.019 | −0.967 | |
| r2 | 0.008 | 0.139 | 0.131 | 0.029 | 0.001 | −0.028 | 0.040 | 0.075 | 0.035 | 0.019 | 0.087 | 0.068 | |
| Bayesian inference | At | 0.967 | 0.885 | −0.082 | 0.951 | 0.934 | −0.017 | 0.934 | 0.918 | −0.016 | 0.984 | 0.885 | −0.099 |
| Av | 0.852 | 0.803 | −0.049 | 0.934 | 0.918 | −0.016 | 0.852 | 0.836 | −0.016 | 0.820 | 0.820 | 0 | |
| Ap | 0.656 | 0.625 | −0.031 | 0.625 | 0.906 | 0.281 | 0.625 | 0.656 | 0.031 | 0.625 | 0.625 | 0 | |
| SVM classification | At | 1.000 | 0.984 | −0.016 | 1.000 | 1.000 | 0 | 1.000 | 1.000 | 0 | 1.000 | 0.984 | −0.016 |
| Av | 0.505 | 0.475 | −0.03 | 0.803 | 0.852 | 0.049 | 0.590 | 0.623 | 0.033 | 0.656 | 0.623 | −0.033 | |
| Ap | 0.656 | 0.719 | 0.063 | 0.875 | 0.875 | 0 | 0.625 | 0.719 | 0.094 | 0.688 | 0.719 | 0.031 | |
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Zhu, R.; Liu, Q.; Tang, J.; Li, H.; Cao, Z. Investigations on Inhibitors of Hedgehog Signal Pathway: A Quantitative Structure-Activity Relationship Study. Int. J. Mol. Sci. 2011, 12, 3018-3033. https://doi.org/10.3390/ijms12053018
Zhu R, Liu Q, Tang J, Li H, Cao Z. Investigations on Inhibitors of Hedgehog Signal Pathway: A Quantitative Structure-Activity Relationship Study. International Journal of Molecular Sciences. 2011; 12(5):3018-3033. https://doi.org/10.3390/ijms12053018
Chicago/Turabian StyleZhu, Ruixin, Qi Liu, Jian Tang, Huiliang Li, and Zhiwei Cao. 2011. "Investigations on Inhibitors of Hedgehog Signal Pathway: A Quantitative Structure-Activity Relationship Study" International Journal of Molecular Sciences 12, no. 5: 3018-3033. https://doi.org/10.3390/ijms12053018





