Claudin 4 Is Differentially Expressed between Ovarian Cancer Subtypes and Plays a Role in Spheroid Formation
Abstract
:1. Introduction
2. Results and Discussion
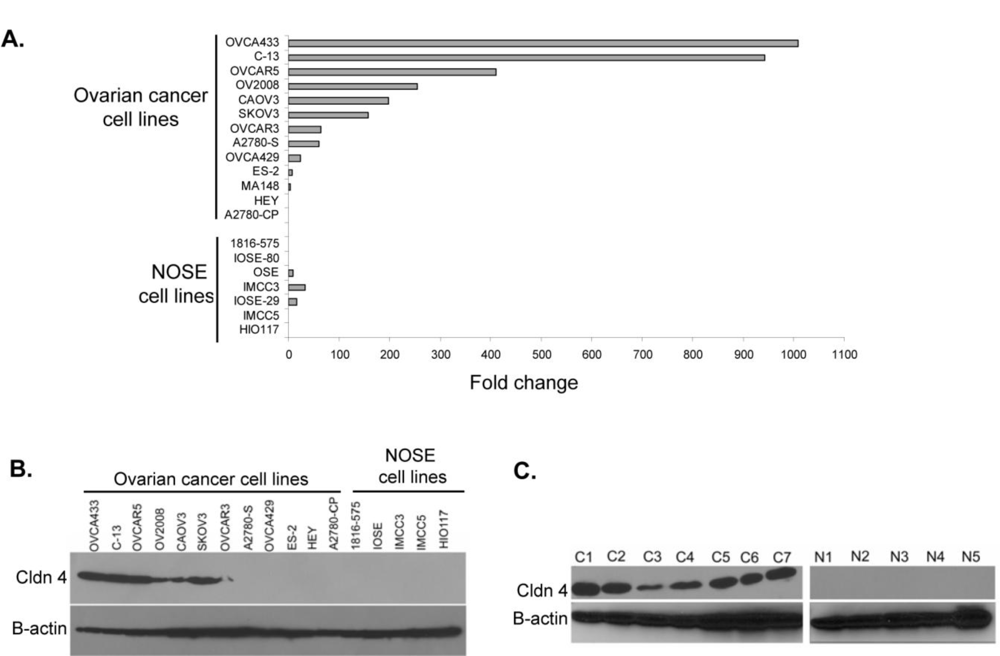
2.1. Claudin 4 RNA Is Overexpressed in Ovarian Cancer
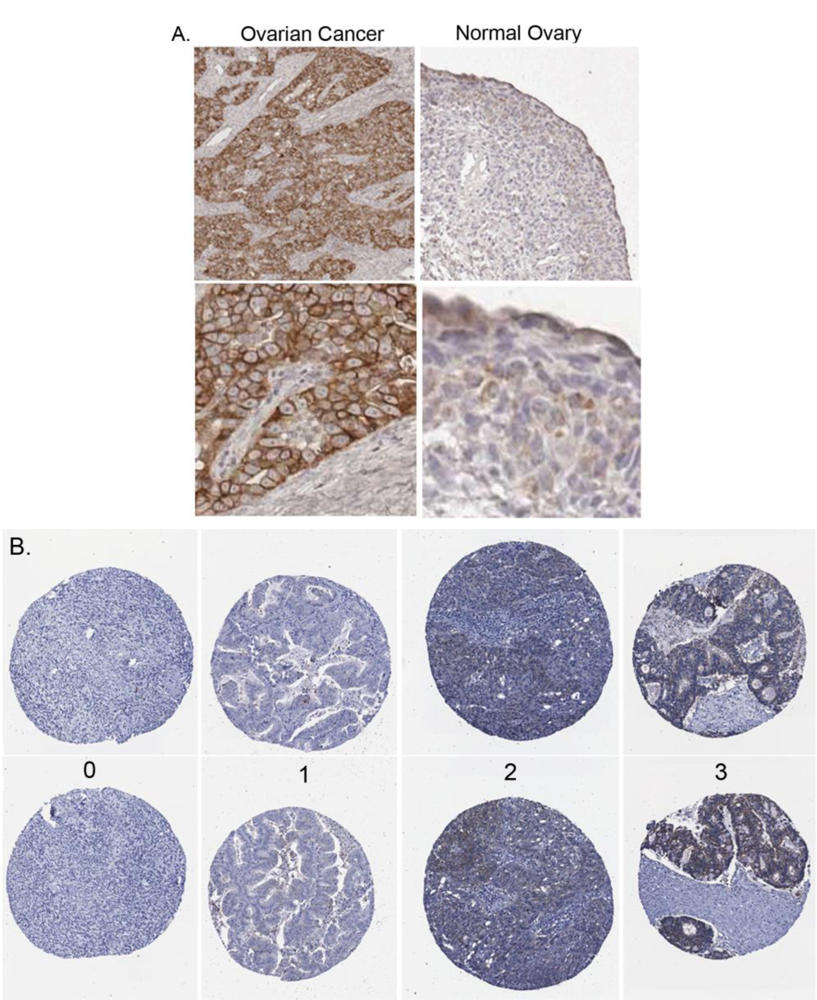
2.2. Claudin 4 Protein Is Overexpressed in Ovarian Cancer Cell Lines and Tissues
2.3. Claudin 4 Protein Expression in Ovarian Cancer Tissues
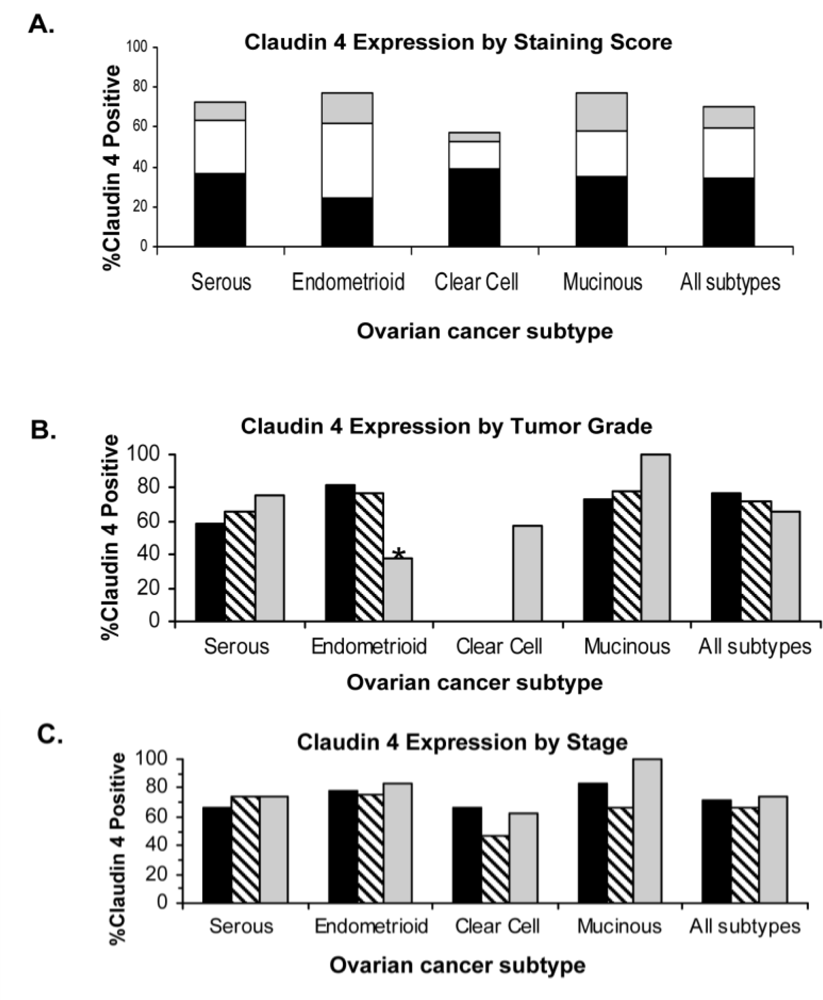
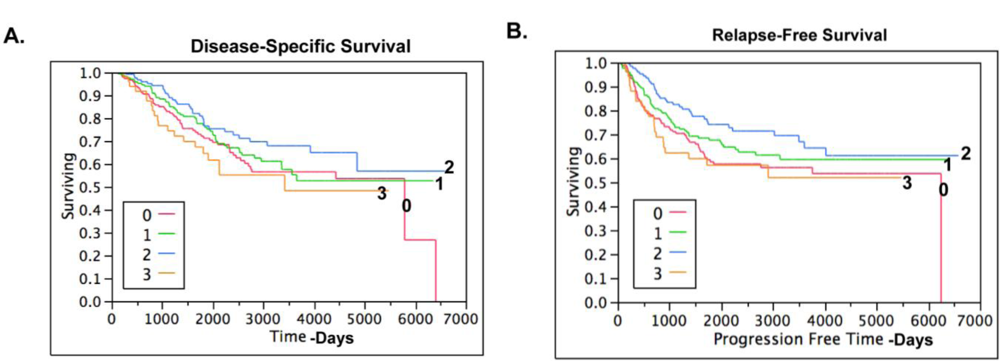
2.4. Claudin 4 Protein Is Differentially Expressed between Subtypes of Ovarian Cancer
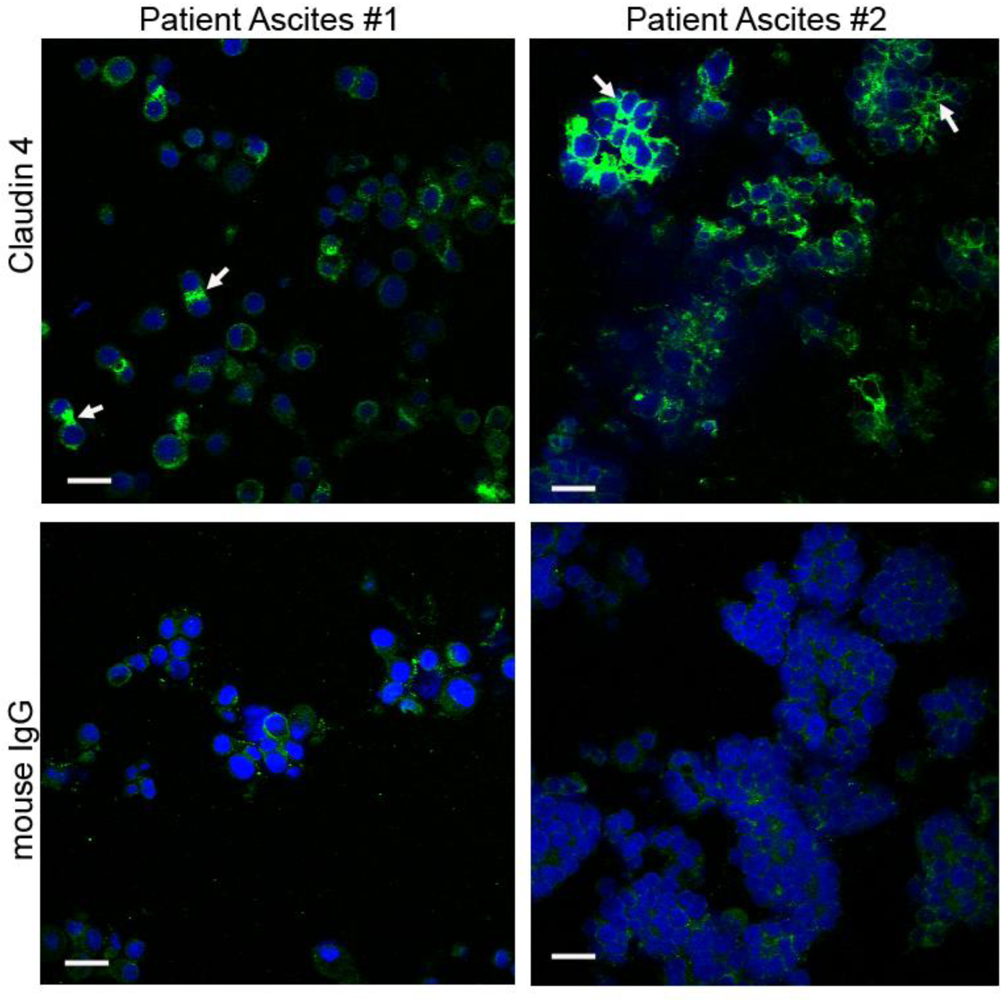
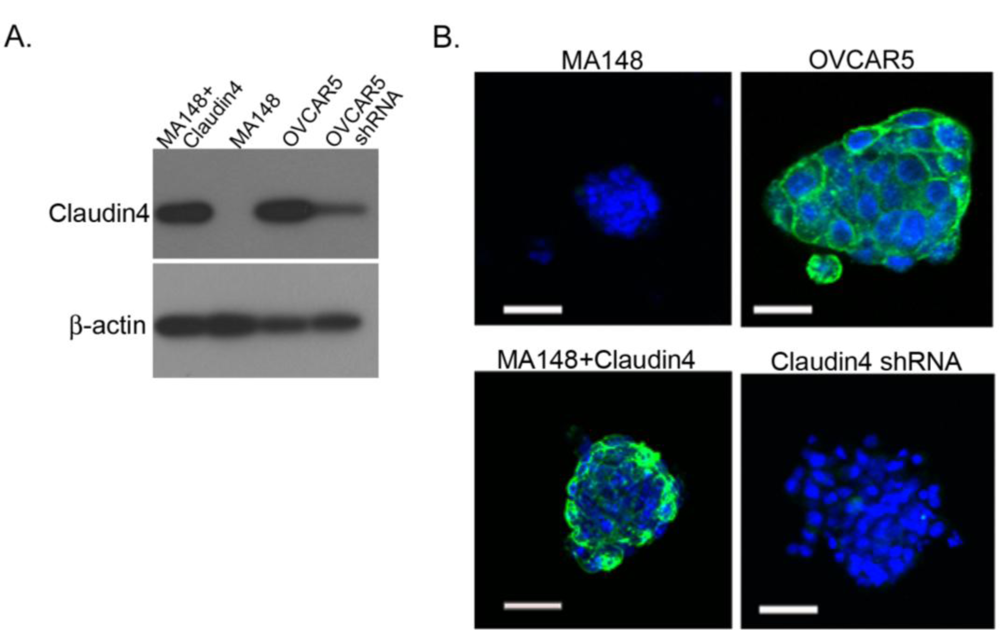
2.5. Claudin 4 Plays a Role in Spheroid Formation/Integrity
2.6. Claudin 4 Increases the Rate of Ovarian Cancer Spheroid Formation
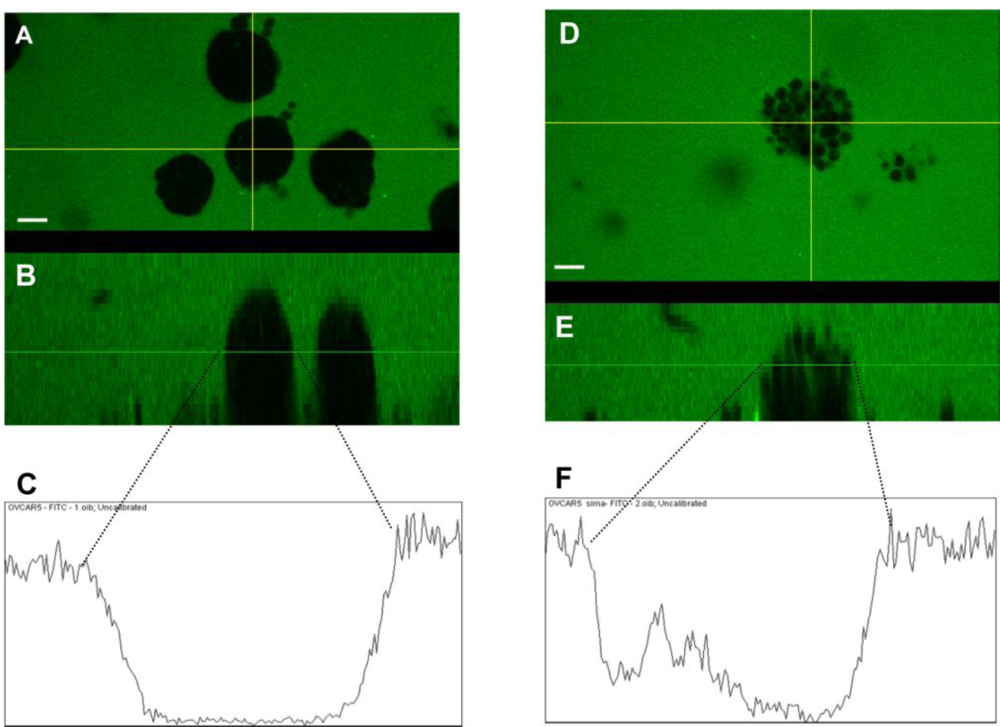
2.7. Claudin 4 Decreases Paracellular Permeability
3. Experimental Section
3.1. Reagents
3.2. Cell Lines
3.3. shRNA Knockdown of Claudin 4
3.4. Transfection of MA148 Cells with Claudin 4
3.5. Tissue Samples
3.6. Isolation of Spheroids from the Ascites of Ovarian Cancer Patients
3.7. RNA Extraction and Reverse Transcriptase Polymerase Chain Reaction
3.8. Quantitative Reverse Transcriptase Polymerase Chain Reaction
3.9. Western Immunoblotting
3.10. Immunohistochemical Staining of Tissues
3.11. Tissue Microarrays
3.12. TMA Statistical Analysis
3.13. Spheroid Formation in Vitro
3.14. Immunocytochemical Staining of Spheroids
3.15. FITC-Dextran Paracellular Permeability Imaging
4. Conclusions
Acknowledgments
References
- Boyle, P; Levin, B. World Cancer Report 2008; International Agency for Research on Cancer: Lyon, France, 2008. [Google Scholar]
- Jemal, A; Siegel, R; Ward, E; Hao, Y; Xu, J; Thun, MJ. Cancer statistics, 2009. CA Cancer J. Clin 2009, 59, 225–249. [Google Scholar]
- Shield, K; Ackland, ML; Ahmed, N; Rice, GE. Multicellular spheroids in ovarian cancer metastases: Biology and pathology. Gynecol. Oncol 2009, 113, 143–148. [Google Scholar]
- Burleson, KM; Boente, MP; Pambuccian, SE; Skubitz, AP. Disaggregation and invasion of ovarian carcinoma ascites spheroids. J. Transl. Med 2006, 4, 6. [Google Scholar]
- Burleson, KM; Casey, RC; Skubitz, KM; Pambuccian, SE; Oegema, TR, Jr; Skubitz, AP. Ovarian carcinoma ascites spheroids adhere to extracellular matrix components and mesothelial cell monolayers. Gynecol. Oncol 2004, 93, 170–181. [Google Scholar]
- Burleson, KM; Hansen, LK; Skubitz, AP. Ovarian carcinoma spheroids disaggregate on type I collagen and invade live human mesothelial cell monolayers. Clin. Exp. Metastasis 2004, 21, 685–697. [Google Scholar]
- Shield, K; Riley, C; Quinn, MA; Rice, GE; Ackland, ML; Ahmed, N. Alpha2beta1 integrin affects metastatic potential of ovarian carcinoma spheroids by supporting disaggregation and proteolysis. J. Carcinog 2007, 6, 11. [Google Scholar]
- Sodek, KL; Ringuette, MJ; Brown, TJ. Compact spheroid formation by ovarian cancer cells is associated with contractile behavior and an invasive phenotype. Int. J. Cancer 2009, 124, 2060–2070. [Google Scholar]
- Desoize, B; Jardillier, J. Multicellular resistance: a paradigm for clinical resistance? Crit. Rev. Oncol. Hematol 2000, 36, 193–207. [Google Scholar]
- Yoshida, Y; Kurokawa, T; Nishikawa, Y; Orisa, M; Kleinman, HK; Kotsuji, F. Laminin-1-derived scrambled peptide AG73T disaggregates laminin-1-induced ovarian cancer cell spheroids and improves the efficacy of cisplatin. Int. J. Oncol 2008, 32, 673–681. [Google Scholar]
- Bignotti, E; Tassi, RA; Calza, S; Ravaggi, A; Romani, C; Rossi, E; Falchetti, M; Odicino, FE; Pecorelli, S; Santin, AD. Differential gene expression profiles between tumor biopsies and short-term primary cultures of ovarian serous carcinomas: identification of novel molecular biomarkers for early diagnosis and therapy. Gynecol. Oncol 2006, 103, 405–416. [Google Scholar]
- Hibbs, K; Skubitz, KM; Pambuccian, SE; Casey, RC; Burleson, KM; Oegema, TR, Jr; Thiele, JJ; Grindle, SM; Bliss, RL; Skubitz, AP. Differential gene expression in ovarian carcinoma: identification of potential biomarkers. Am. J. Pathol 2004, 165, 397–414. [Google Scholar]
- Hough, CD; Sherman-Baust, CA; Pizer, ES; Montz, FJ; Im, DD; Rosenshein, NB; Cho, KR; Riggins, GJ; Morin, PJ. Large-scale serial analysis of gene expression reveals genes differentially expressed in ovarian cancer. Cancer Res 2000, 60, 6281–6287. [Google Scholar]
- Lu, KH; Patterson, AP; Wang, L; Marquez, RT; Atkinson, EN; Baggerly, KA; Ramoth, LR; Rosen, DG; Liu, J; Hellstrom, I; Smith, D; Hartmann, L; Fishman, D; Berchuck, A; Schmandt, R; Whitaker, R; Gershenson, DM; Mills, GB; Bast, RC, Jr. Selection of potential markers for epithelial ovarian cancer with gene expression arrays and recursive descent partition analysis. Clin. Cancer Res 2004, 10, 3291–3300. [Google Scholar]
- Santin, AD; Zhan, F; Bellone, S; Palmieri, M; Cane, S; Bignotti, E; Anfossi, S; Gokden, M; Dunn, D; Roman, JJ; O’Brien, TJ; Tian, E; Cannon, MJ; Shaughnessy, J, Jr; Pecorelli, S. Gene expression profiles in primary ovarian serous papillary tumors and normal ovarian epithelium: identification of candidate molecular markers for ovarian cancer diagnosis and therapy. Int. J. Cancer 2004, 112, 14–25. [Google Scholar]
- Morin, PJ. Claudin proteins in human cancer: promising new targets for diagnosis and therapy. Cancer Res 2005, 65, 9603–9606. [Google Scholar]
- Singh, AB; Sharma, A; Dhawan, P. Claudin family of proteins and cancer: an overview. J. Oncol 2010, 2010, 541957. [Google Scholar]
- Cohn, ML; Goncharuk, VN; Diwan, AH; Zhang, PS; Shen, SS; Prieto, VG. Loss of claudin-1 expression in tumor-associated vessels correlates with acquisition of metastatic phenotype in melanocytic neoplasms. J. Cutan. Pathol 2005, 32, 533–536. [Google Scholar]
- Kim, TH; Huh, JH; Lee, S; Kang, H; Kim, GI; An, HJ. Down-regulation of claudin-2 in breast carcinomas is associated with advanced disease. Histopathology 2008, 53, 48–55. [Google Scholar]
- Miyamoto, K; Kusumi, T; Sato, F; Kawasaki, H; Shibata, S; Ohashi, M; Hakamada, K; Sasaki, M; Kijima, H. Decreased expression of claudin-1 is correlated with recurrence status in esophageal squamous cell carcinoma. Biomed. Res 2008, 29, 71–76. [Google Scholar]
- Morohashi, S; Kusumi, T; Sato, F; Odagiri, H; Chiba, H; Yoshihara, S; Hakamada, K; Sasaki, M; Kijima, H. Decreased expression of claudin-1 correlates with recurrence status in breast cancer. Int. J. Mol. Med 2007, 20, 139–143. [Google Scholar]
- Ohtani, S; Terashima, M; Satoh, J; Soeta, N; Saze, Z; Kashimura, S; Ohsuka, F; Hoshino, Y; Kogure, M; Gotoh, M. Expression of tight-junction-associated proteins in human gastric cancer: downregulation of claudin-4 correlates with tumor aggressiveness and survival. Gastric. Cancer 2009, 12, 43–51. [Google Scholar]
- Oshima, T; Kunisaki, C; Yoshihara, K; Yamada, R; Yamamoto, N; Sato, T; Makino, H; Yamagishi, S; Nagano, Y; Fujii, S; Shiozawa, M; Akaike, M; Wada, N; Rino, Y; Masuda, M; Tanaka, K; Imada, T. Reduced expression of the claudin-7 gene correlates with venous invasion and liver metastasis in colorectal cancer. Oncol. Rep 2008, 19, 953–959. [Google Scholar]
- Sauer, T; Pedersen, MK; Ebeltoft, K; Naess, O. Reduced expression of Claudin-7 in fine needle aspirates from breast carcinomas correlate with grading and metastatic disease. Cytopathology 2005, 16, 193–198. [Google Scholar]
- Sung, CO; Han, SY; Kim, SH. Low Expression of Claudin-4 is Associated with Poor Prognosis in Esophageal Squamous Cell Carcinoma. Ann. Surg. Oncol 2011, 18, 273–281. [Google Scholar]
- Ueda, J; Semba, S; Chiba, H; Sawada, N; Seo, Y; Kasuga, M; Yokozaki, H. Heterogeneous expression of claudin-4 in human colorectal cancer: decreased claudin-4 expression at the invasive front correlates cancer invasion and metastasis. Pathobiology 2007, 74, 32–41. [Google Scholar]
- Usami, Y; Chiba, H; Nakayama, F; Ueda, J; Matsuda, Y; Sawada, N; Komori, T; Ito, A; Yokozaki, H. Reduced expression of claudin-7 correlates with invasion and metastasis in squamous cell carcinoma of the esophagus. Hum. Pathol 2006, 37, 569–577. [Google Scholar]
- Agarwal, R; D’Souza, T; Morin, PJ. Claudin-3 and claudin-4 expression in ovarian epithelial cells enhances invasion and is associated with increased matrix metalloproteinase-2 activity. Cancer Res 2005, 65, 7378–7385. [Google Scholar]
- Landers, KA; Samaratunga, H; Teng, L; Buck, M; Burger, MJ; Scells, B; Lavin, MF; Gardiner, RA. Identification of claudin-4 as a marker highly overexpressed in both primary and metastatic prostate cancer. Br. J. Cancer 2008, 99, 491–501. [Google Scholar]
- Lanigan, F; McKiernan, E; Brennan, DJ; Hegarty, S; Millikan, RC; McBryan, J; Jirstrom, K; Landberg, G; Martin, F; Duffy, MJ; Gallagher, WM. Increased claudin-4 expression is associated with poor prognosis and high tumour grade in breast cancer. Int. J. Cancer 2009, 124, 2088–2097. [Google Scholar]
- Long, H; Crean, CD; Lee, WH; Cummings, OW; Gabig, TG. Expression of Clostridium perfringens enterotoxin receptors claudin-3 and claudin-4 in prostate cancer epithelium. Cancer Res 2001, 61, 7878–7881. [Google Scholar]
- Rangel, LB; Agarwal, R; D’Souza, T; Pizer, ES; Alo, PL; Lancaster, WD; Gregoire, L; Schwartz, DR; Cho, KR; Morin, PJ. Tight junction proteins claudin-3 and claudin-4 are frequently overexpressed in ovarian cancer but not in ovarian cystadenomas. Clin. Cancer Res 2003, 9, 2567–2575. [Google Scholar]
- Takehara, M; Nishimura, T; Mima, S; Hoshino, T; Mizushima, T. Effect of claudin expression on paracellular permeability, migration and invasion of colonic cancer cells. Biol. Pharm. Bull 2009, 32, 825–831. [Google Scholar]
- Morita, K; Tsukita, S; Miyachi, Y. Tight junction-associated proteins (occludin, ZO-1, claudin-1, claudin-4) in squamous cell carcinoma and Bowen’s disease. Br. J. Dermatol 2004, 151, 328–334. [Google Scholar]
- Sato, N; Fukushima, N; Maitra, A; Iacobuzio-Donahue, CA; van Heek, NT; Cameron, JL; Yeo, CJ; Hruban, RH; Goggins, M. Gene expression profiling identifies genes associated with invasive intraductal papillary mucinous neoplasms of the pancreas. Am. J. Pathol 2004, 164, 903–914. [Google Scholar]
- Gress, TM; Muller-Pillasch, F; Geng, M; Zimmerhackl, F; Zehetner, G; Friess, H; Buchler, M; Adler, G; Lehrach, H. A pancreatic cancer-specific expression profile. Oncogene 1996, 13, 1819–1830. [Google Scholar]
- Litkouhi, B; Kwong, J; Lo, CM; Smedley, JG, 3rd; McClane, BA; Aponte, M; Gao, Z; Sarno, JL; Hinners, J; Welch, WR; Berkowitz, RS; Mok, SC; Garner, EI. Claudin-4 overexpression in epithelial ovarian cancer is associated with hypomethylation and is a potential target for modulation of tight junction barrier function using a C-terminal fragment of Clostridium perfringens enterotoxin. Neoplasia 2007, 9, 304–314. [Google Scholar]
- Zhu, Y; Brannstrom, M; Janson, PO; Sundfeldt, K. Differences in expression patterns of the tight junction proteins,claudin 1, 3, 4 and 5, in human ovarian surface epithelium as compared to epithelia in inclusion cysts and epithelial ovarian tumours. Int. J. Cancer 2006, 118, 1884–1891. [Google Scholar]
- Crum, CP; Drapkin, R; Miron, A; Ince, TA; Muto, M; Kindelberger, DW; Lee, Y. The distal fallopian tube: a new model for pelvic serous carcinogenesis. Curr. Opin. Obstet. Gynecol 2007, 19, 3–9. [Google Scholar]
- Gilks, CB; Ionescu, DN; Kalloger, SE; Kobel, M; Irving, J; Clarke, B; Santos, J; Le, N; Moravan, V; Swenerton, K. Tumor cell type can be reproducibly diagnosed and is of independent prognostic significance in patients with maximally debulked ovarian carcinoma. Hum. Pathol 2008, 39, 1239–1251. [Google Scholar]
- Soini, Y; Talvensaari-Mattila, A. Expression of claudins 1, 4, 5, and 7 in ovarian tumors of diverse types. Int. J. Gynecol. Pathol 2006, 25, 330–335. [Google Scholar]
- Lechpammer, M; Resnick, MB; Sabo, E; Yakirevich, E; Greaves, WO; Sciandra, KT; Tavares, R; Noble, LC; DeLellis, RA; Wang, LJ. The diagnostic and prognostic utility of claudin expression in renal cell neoplasms. Mod. Pathol 2008, 21, 1320–1329. [Google Scholar]
- Kleinberg, L; Holth, A; Trope, CG; Reich, R; Davidson, B. Claudin upregulation in ovarian carcinoma effusions is associated with poor survival. Hum. Pathol 2008, 39, 747–757. [Google Scholar]
- Ivascu, A; Kubbies, M. Diversity of cell-mediated adhesions in breast cancer spheroids. Int. J. Oncol 2007, 31, 1403–1413. [Google Scholar]
- Bates, RC; Edwards, NS; Yates, JD. Spheroids and cell survival. Crit. Rev. Oncol. Hematol 2000, 36, 61–74. [Google Scholar]
- Frankel, A; Rosen, K; Filmus, J; Kerbel, RS. Induction of anoikis and suppression of human ovarian tumor growth in vivo by down-regulation of Bcl-X(L). Cancer Res 2001, 61, 4837–4841. [Google Scholar]
- Makhija, S; Taylor, DD; Gibb, RK; Gercel-Taylor, C. Taxol-induced bcl-2 phosphorylation in ovarian cancer cell monolayer and spheroids. Int. J. Oncol 1999, 14, 515–521. [Google Scholar]
- Stewart, JJ; White, JT; Yan, X; Collins, S; Drescher, CW; Urban, ND; Hood, L; Lin, B. Proteins associated with Cisplatin resistance in ovarian cancer cells identified by quantitative proteomic technology and integrated with mRNA expression levels. Mol. Cell. Proteomics 2006, 5, 433–443. [Google Scholar]
- Heinzelmann-Schwarz, VA; Gardiner-Garden, M; Henshall, SM; Scurry, J; Scolyer, RA; Davies, MJ; Heinzelmann, M; Kalish, LH; Bali, A; Kench, JG; Edwards, LS; Vanden Bergh, PM; Hacker, NF; Sutherland, RL; O’Brien, PM. Overexpression of the cell adhesion molecules DDR1, Claudin 3, and Ep-CAM in metaplastic ovarian epithelium and ovarian cancer. Clin. Cancer Res 2004, 10, 4427–4436. [Google Scholar]
- D’Souza, T; Indig, FE; Morin, PJ. Phosphorylation of claudin-4 by PKCepsilon regulates tight junction barrier function in ovarian cancer cells. Exp. Cell Res 2007, 313, 3364–3375. [Google Scholar]
- Tanaka, M; Kamata, R; Sakai, R. EphA2 phosphorylates the cytoplasmic tail of Claudin-4 and mediates paracellular permeability. J. Biol. Chem 2005, 280, 42375–42382. [Google Scholar]
- Casey, RC; Burleson, KM; Skubitz, KM; Pambuccian, SE; Oegema, TR, Jr; Ruff, LE; Skubitz, AP. Beta 1-integrins regulate the formation and adhesion of ovarian carcinoma multicellular spheroids. Am. J. Pathol 2001, 159, 2071–2080. [Google Scholar]
- Casey, RC; Koch, KA; Oegema, TR, Jr; Skubitz, KM; Pambuccian, SE; Grindle, SM; Skubitz, AP. Establishment of an in vitro assay to measure the invasion of ovarian carcinoma cells through mesothelial cell monolayers. Clin. Exp. Metastasis 2003, 20, 343–356. [Google Scholar]
- L’Esperance, S; Bachvarova, M; Tetu, B; Mes-Masson, AM; Bachvarov, D. Global gene expression analysis of early response to chemotherapy treatment in ovarian cancer spheroids. BMC Genomics 2008, 9, 99. [Google Scholar]
- Katahira, J; Inoue, N; Horiguchi, Y; Matsuda, M; Sugimoto, N. Molecular cloning and functional characterization of the receptor for Clostridium perfringens enterotoxin. J. Cell. Biol 1997, 136, 1239–1247. [Google Scholar]
- McClane, BA. Clostridium perfringens enterotoxin acts by producing small molecule permeability alterations in plasma membranes. Toxicology 1994, 87, 43–67. [Google Scholar]
- Moriwaki, K; Tsukita, S; Furuse, M. Tight junctions containing claudin 4 and 6 are essential for blastocyst formation in preimplantation mouse embryos. Dev. Biol 2007, 312, 509–522. [Google Scholar]
- Kondoh, M; Masuyama, A; Takahashi, A; Asano, N; Mizuguchi, H; Koizumi, N; Fujii, M; Hayakawa, T; Horiguchi, Y; Watanbe, Y. A novel strategy for the enhancement of drug absorption using a claudin modulator. Mol. Pharmacol 2005, 67, 749–756. [Google Scholar]
- Shaw, TJ; Senterman, MK; Dawson, K; Crane, CA; Vanderhyden, BC. Characterization of intraperitoneal, orthotopic, and metastatic xenograft models of human ovarian cancer. Mol. Ther 2004, 10, 1032–1042. [Google Scholar]
- Subramanian, IV; Bui Nguyen, TM; Truskinovsky, AM; Tolar, J; Blazar, BR; Ramakrishnan, S. Adeno-associated virus-mediated delivery of a mutant endostatin in combination with carboplatin treatment inhibits orthotopic growth of ovarian cancer and improves long-term survival. Cancer Res 2006, 66, 4319–4328. [Google Scholar]
- Skubitz, AP; Campbell, KD; Goueli, S; Skubitz, KM. Association of beta 1 integrin with protein kinase activity in large detergent resistant complexes. FEBS Lett 1998, 426, 386–391. [Google Scholar]
- Kruk, PA; Maines-Bandiera, SL; Auersperg, N. A simplified method to culture human ovarian surface epithelium. Lab. Invest 1990, 63, 132–136. [Google Scholar]
- Nicosia, SV; Wilbanks, GD; Saunders, B; Mayer, J; Cardosi, RJ; Kruk, PA; Cheng, J; Bai, W; Coppola, D; Fiorica, J. Cytology of human ovarian surface epithelial brushings. Cancer 2004, 102, 1–10. [Google Scholar]
- Fronhoffs, S; Totzke, G; Stier, S; Wernert, N; Rothe, M; Bruning, T; Koch, B; Sachinidis, A; Vetter, H; Ko, Y. A method for the rapid construction of cRNA standard curves in quantitative real-time reverse transcription polymerase chain reaction. Mol. Cell. Probes 2002, 16, 99–110. [Google Scholar]
- Andersen, JD; Boylan, KL; Xue, FS; Anderson, LB; Witthuhn, BA; Markowski, TW; Higgins, L; Skubitz, AP. Identification of candidate biomarkers in ovarian cancer serum by depletion of highly abundant proteins and differential in-gel electrophoresis. Electrophoresis 2010, 31, 599–610. [Google Scholar]
- DeRycke, MS; Andersen, JD; Harrington, KM; Pambuccian, SE; Kalloger, SE; Boylan, KL; Argenta, PA; Skubitz, AP. S100A1 expression in ovarian and endometrial endometrioid carcinomas is a prognostic indicator of relapse-free survival. Am. J. Clin. Pathol 2009, 132, 846–856. [Google Scholar]
- McCluggage, WG. My approach to and thoughts on the typing of ovarian carcinomas. J. Clin. Pathol 2008, 61, 152–163. [Google Scholar]
- Soslow, RA. Histologic subtypes of ovarian carcinoma: an overview. Int. J. Gynecol. Pathol 2008, 27, 161–174. [Google Scholar]
- Silverberg, SG. Histopathologic grading of ovarian carcinoma: a review and proposal. Int. J. Gynecol. Pathol 2000, 19, 7–15. [Google Scholar]
- Cocco, E; Casagrande, F; Bellone, S; Richter, CE; Bellone, M; Todeschini, P; Holmberg, JC; Fu, HH; Montagna, MK; Mor, G; Schwartz, PE; Arin-Silasi, D; Azoudi, M; Rutherford, TJ; Abu-Khalaf, M; Pecorelli, S; Santin, AD. Clostridium perfringens enterotoxin carboxy-terminal fragment is a novel tumor-homing peptide for human ovarian cancer. BMC Cancer 2010, 10, 349. [Google Scholar]
- Kakutani, H; Kondoh, M; Saeki, R; Fujii, M; Watanabe, Y; Mizuguchi, H; Yagi, K. Claudin-4-targeting of diphtheria toxin fragment A using a C-terminal fragment of Clostridium perfringens enterotoxin. Eur. J. Pharm. Biopharm 2010, 75, 213–217. [Google Scholar]
- Ebihara, C; Kondoh, M; Hasuike, N; Harada, M; Mizuguchi, H; Horiguchi, Y; Fujii, M; Watanabe, Y. Preparation of a claudin-targeting molecule using a C-terminal fragment of Clostridium perfringens enterotoxin. J. Pharmacol. Exp. Ther 2006, 316, 255–260. [Google Scholar]
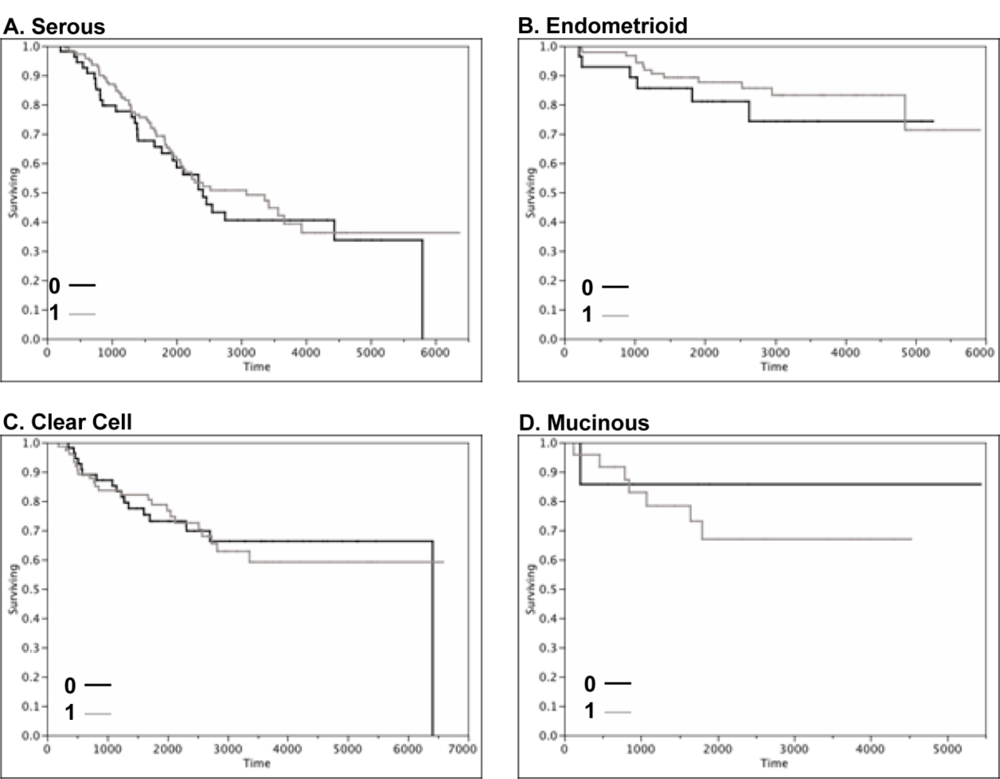

| Subtype | Median Age (range) | Stage | N | Claudin 4 Positive (%) | Silverberg Grade | N | Claudin 4 Positive (%) | Claudin 4 Positive Overall (%) |
|---|---|---|---|---|---|---|---|---|
| Serous (n = 212) | 59.6 (33.5–86.0) | I | 50 | 33 (66.0%) | 1 | 12 | 7 (58.3 %) | 153 (72.2 %) |
| II | 93 | 69 (74.2%) | 2 | 56 | 37 (66.1%) | |||
| III | 69 | 51 (73.9%) | 3 | 144 | 109 (75.7%) | |||
| Endometrioid (n = 125) | 54.1 (29.4–88.1) | I | 69 | 54 (78.3%) | 1 | 82 | 66 (81.5%) | 96 (77.4 %) |
| II | 50 | 37 (75.5%) | 2 | 35 | 27 (77.1%) | |||
| III | 6 | 5 (83.3%) | 3 | 8 | 3 (37.5%) | |||
| Clear Cell (n = 132) | 55.0 (28.1–89.0) | I | 68 | 45 (66.3%) | 1 | 0* | N/A | 76 (57.6 %) |
| II | 56 | 26 (46.4%) | 2 | 0* | N/A | |||
| III | 8 | 5 (62.5%) | 3 | 132 | 76 (57.6 %) | |||
| Mucinous (n = 31) | 56.4 (25.4–76.7) | I | 18 | 15 (83.3%) | 1 | 11 | 8 (72.7%) | 24 (77.4%) |
| II | 12 | 8 (66.7%) | 2 | 18 | 14 (77.8%) | |||
| III | 1 | 1 (100%) | 3 | 2 | 2 (100.0%) | |||
| Total (n = 500) | 56.6 (25.4–89.0) | I | 205 | 147 (71.7%) | 1 | 105 | 81 (77.1%) | 349 (69.9%) |
| II | 211 | 140 (66.4%) | 2 | 109 | 78 (71.6%) | |||
| III | 84 | 62 (73.8%) | 3 | 286 | 190 (66.4%) | |||
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Boylan, K.L.M.; Misemer, B.; DeRycke, M.S.; Andersen, J.D.; Harrington, K.M.; Kalloger, S.E.; Gilks, C.B.; Pambuccian, S.E.; Skubitz, A.P.N. Claudin 4 Is Differentially Expressed between Ovarian Cancer Subtypes and Plays a Role in Spheroid Formation. Int. J. Mol. Sci. 2011, 12, 1334-1358. https://doi.org/10.3390/ijms12021334
Boylan KLM, Misemer B, DeRycke MS, Andersen JD, Harrington KM, Kalloger SE, Gilks CB, Pambuccian SE, Skubitz APN. Claudin 4 Is Differentially Expressed between Ovarian Cancer Subtypes and Plays a Role in Spheroid Formation. International Journal of Molecular Sciences. 2011; 12(2):1334-1358. https://doi.org/10.3390/ijms12021334
Chicago/Turabian StyleBoylan, Kristin L. M., Benjamin Misemer, Melissa S. DeRycke, John D. Andersen, Katherine M. Harrington, Steve E. Kalloger, C. Blake Gilks, Stefan E. Pambuccian, and Amy P. N. Skubitz. 2011. "Claudin 4 Is Differentially Expressed between Ovarian Cancer Subtypes and Plays a Role in Spheroid Formation" International Journal of Molecular Sciences 12, no. 2: 1334-1358. https://doi.org/10.3390/ijms12021334





