Use of Oligonucleotides Carrying Photolabile Groups for the Control of the Deposition of Nanoparticles in Surfaces and Nanoparticle Association
Abstract
:1. Introduction
2. Results and Discussion
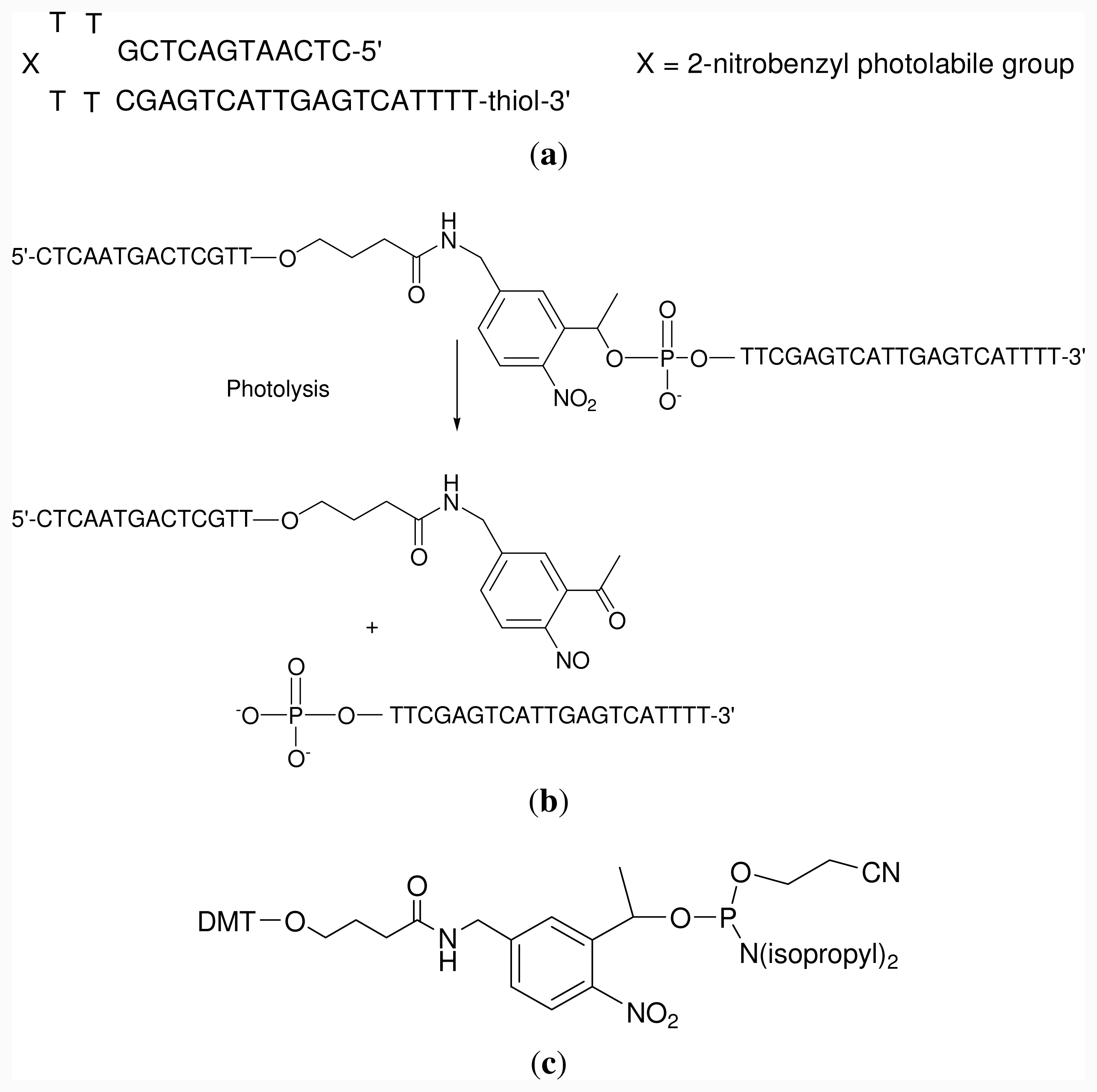
2.1. Oligonucleotide Synthesis
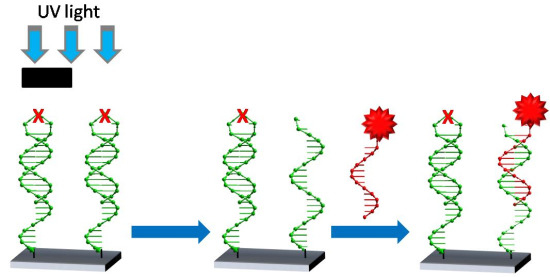
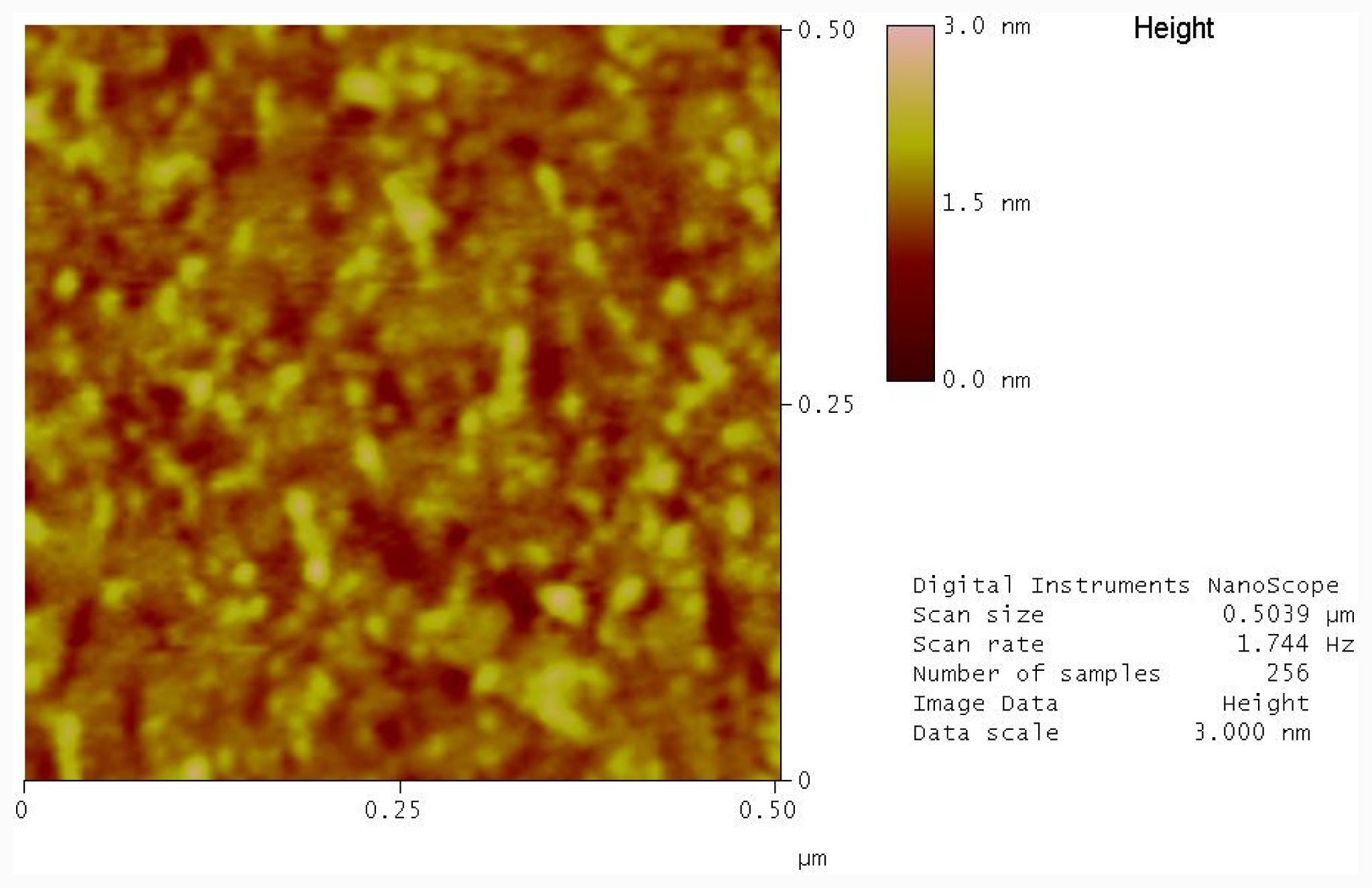
2.2. Immobilization of Photocleavable Hairpin to Gold Substrates
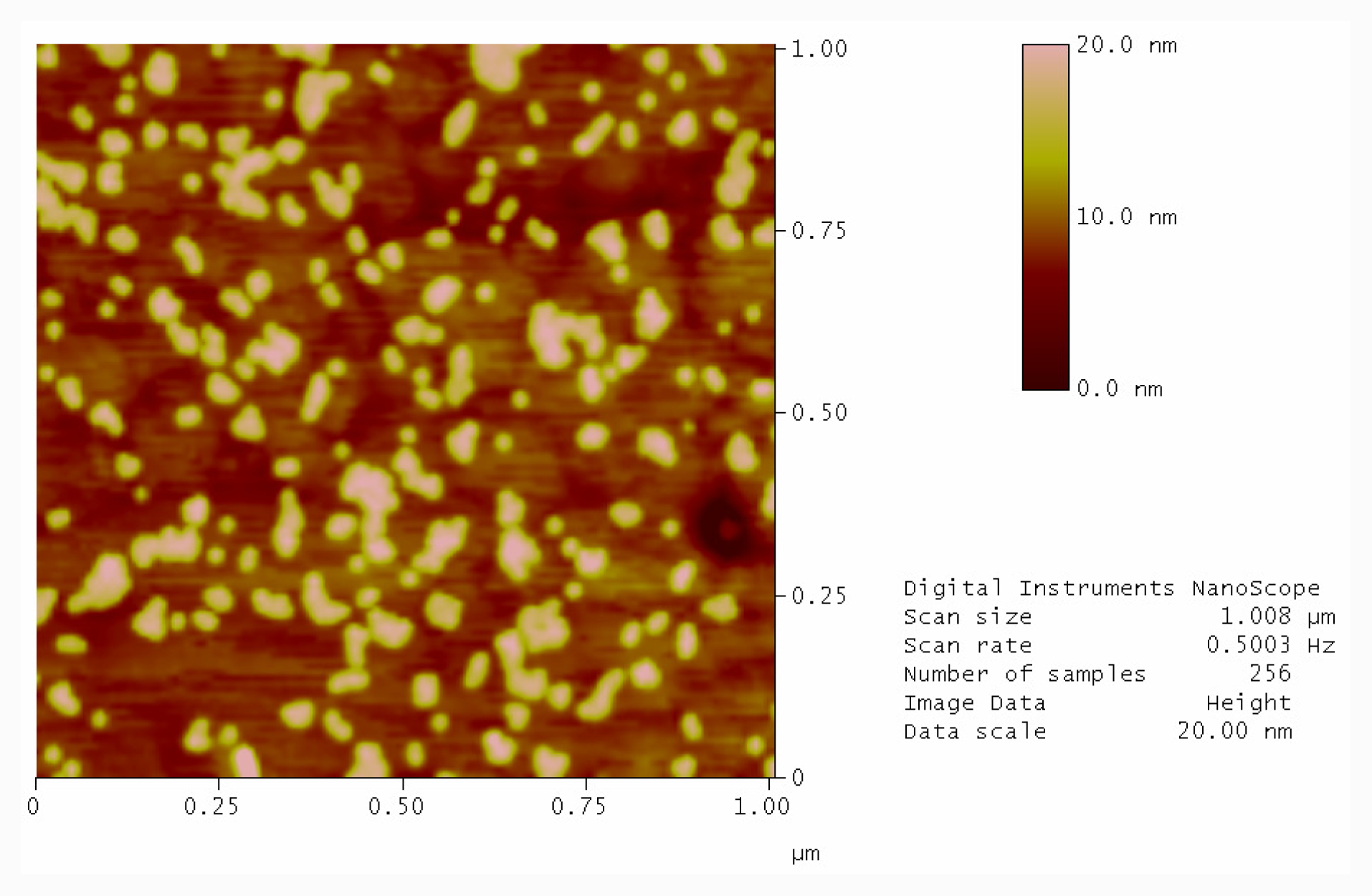
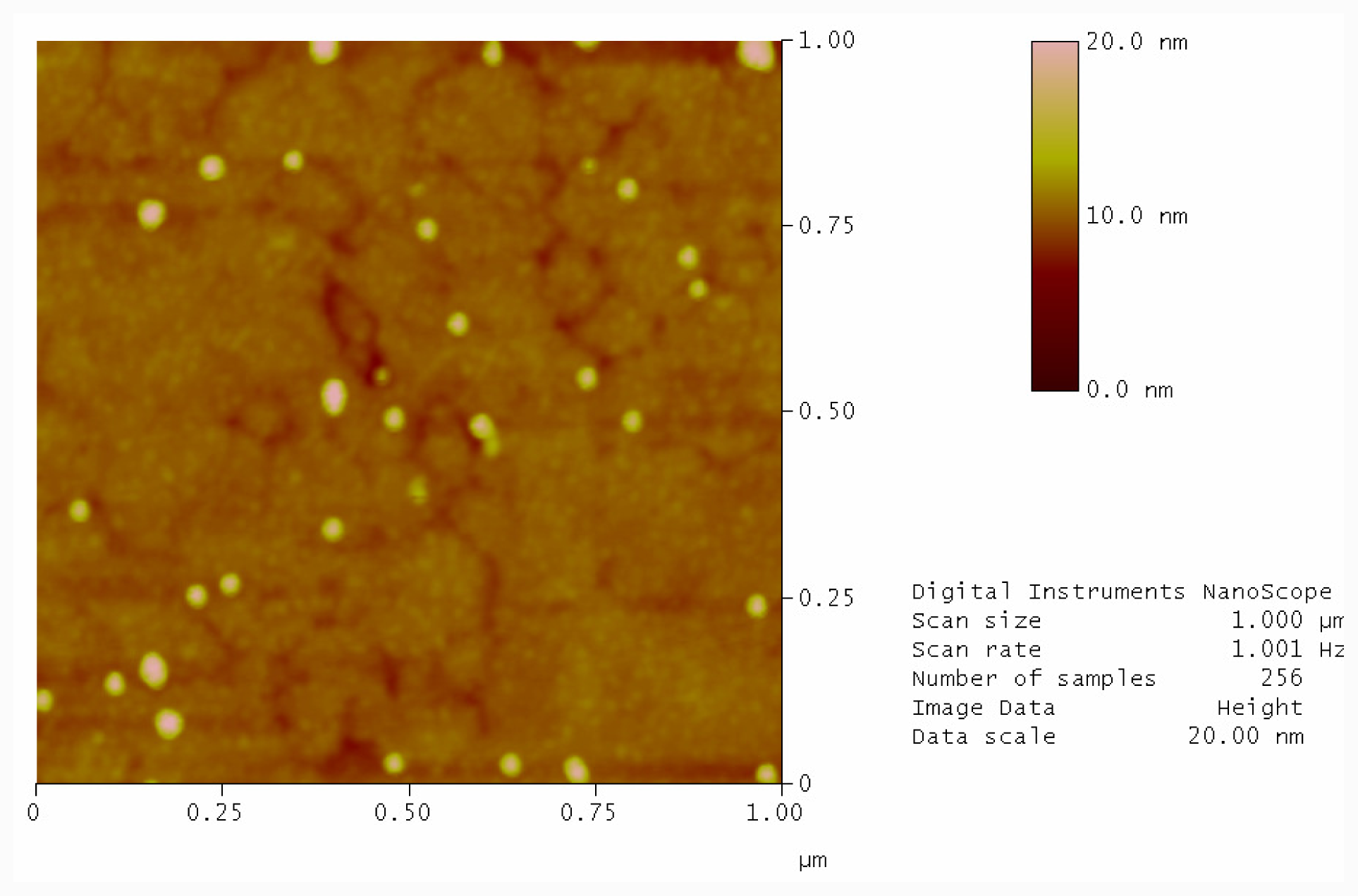
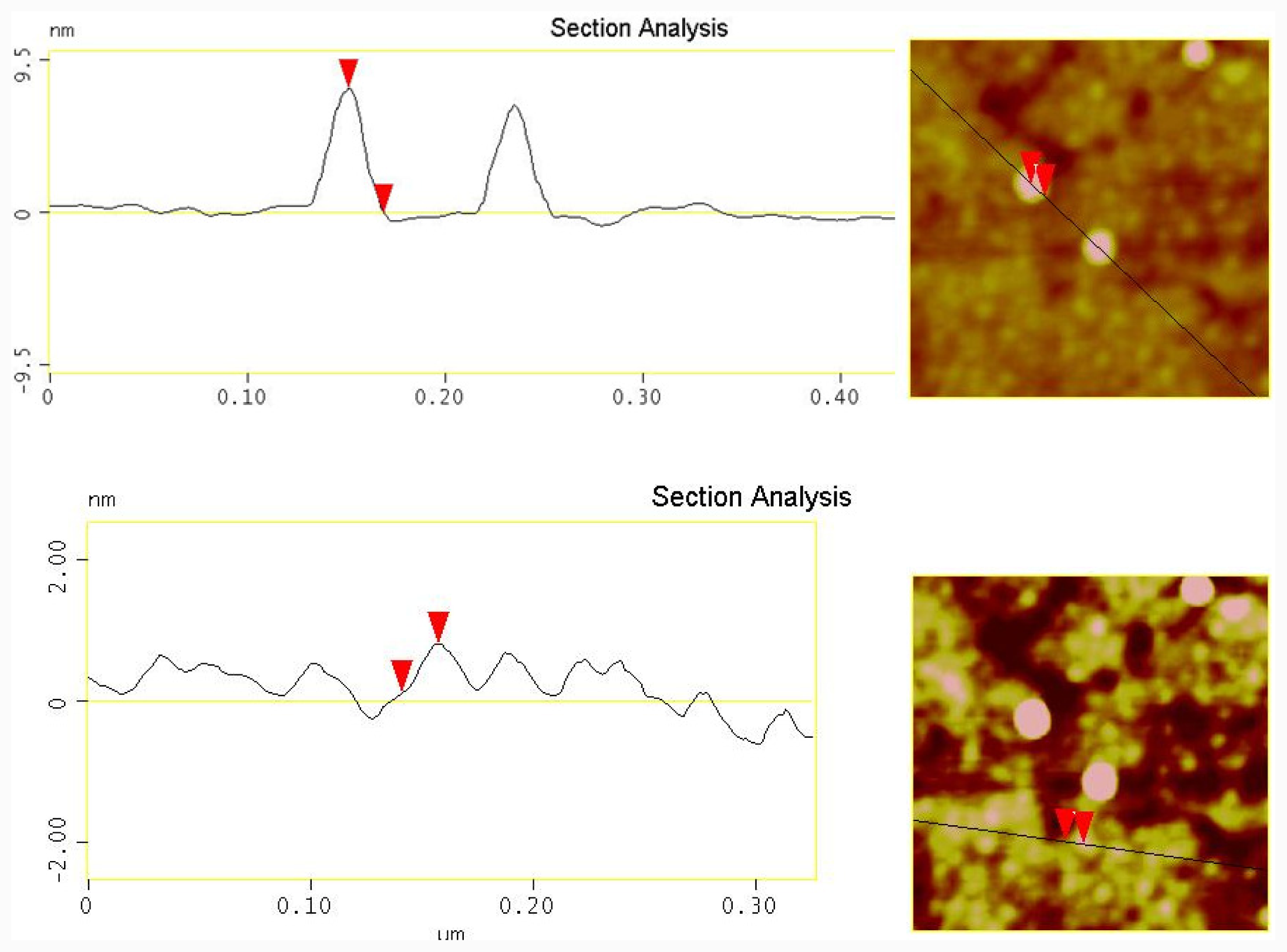
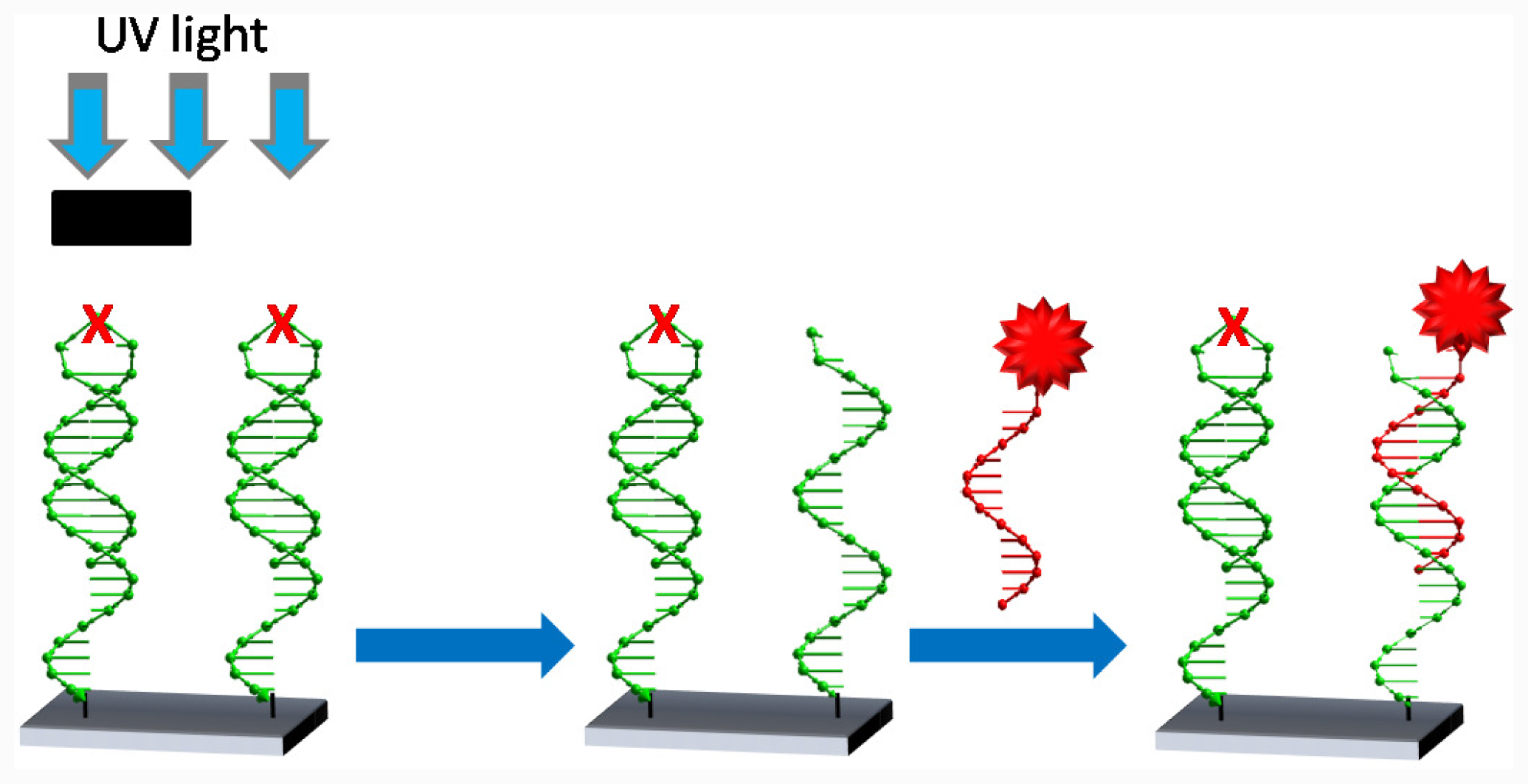
2.3. Photolysis of Hairpin Oligonucleotide A on a Template Stripped Gold Substrate
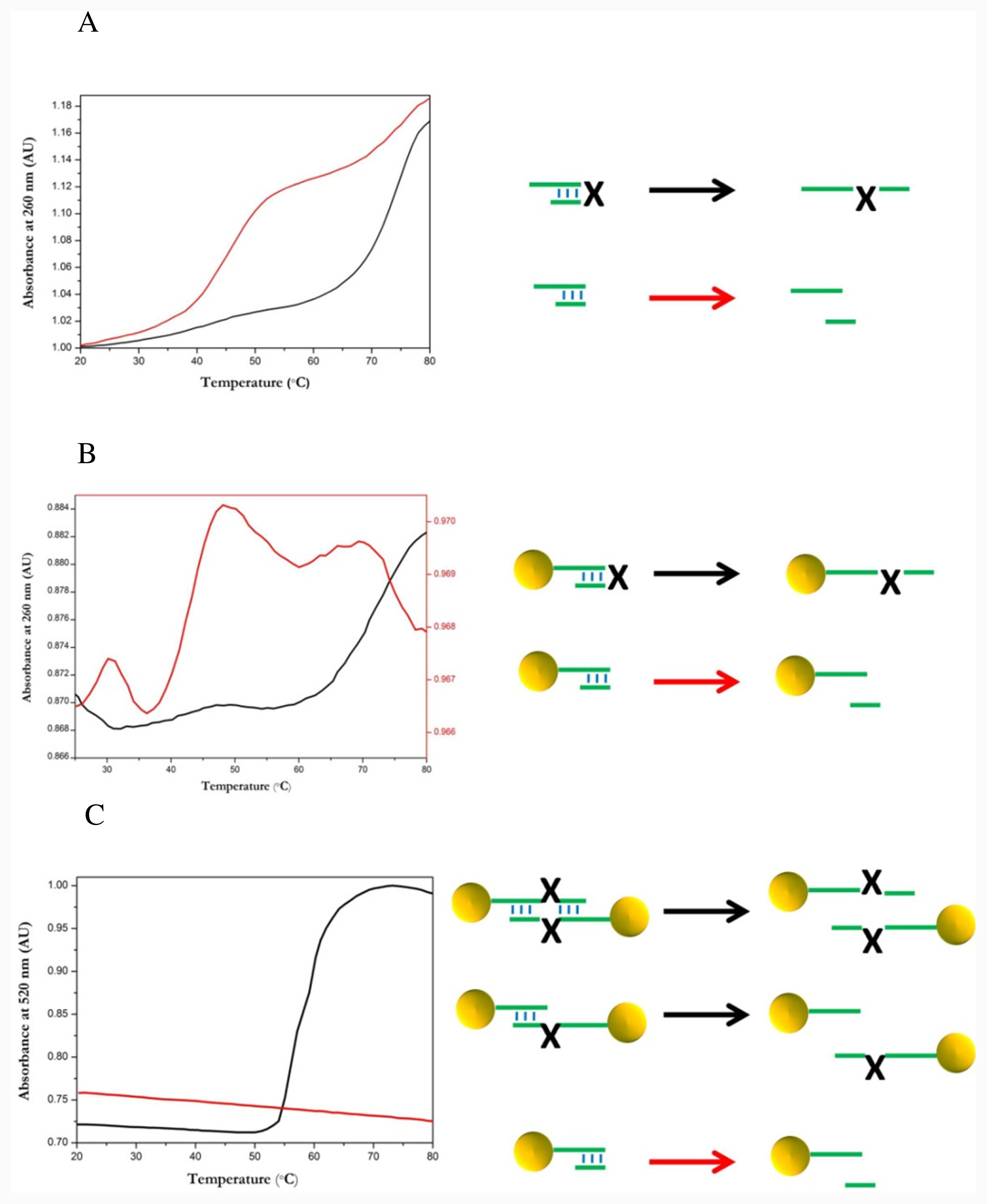
2.4. Thermal Dissociation Analysis Hairpin Oligonucleotide A Linked to Gold Nanoparticles
3. Experimental Section
3.1. General
3.2. Oligodeoxynucleotide Synthesis
3.3. Conjugation of Thiolated Oligonucleotides to 10 nm Gold Nanoparticles
3.4. Immobilisation of Hairpin Oligonucleotide A to a Template Stripped Gold Substrate
3.5. Photolysis with Mercury Lamp and Development of Photocleaved Areas with Complementary Oligonucleotide Labelled with Gold Nanoparticles
3.6. Atomic Force Microscopy (AFM)
3.7. Melting Curves of Hairpin Gold Nanoparticle Conjugates
4. Conclusions
Supplementary Material
ijms-12-07238-s001.pdfAcknowledgments
References
- Mirkin, CA; Letsinger, RL; Mucic, RC; Storhoff, JJ. A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature 1996, 382, 607–609. [Google Scholar]
- Park, SJ; Taton, TA; Mirkin, CA. Array-based electrical detection of DNA with nanoparticle probes. Science 2002, 295, 1503–1506. [Google Scholar]
- Taton, TA; Mirkin, CA; Letsinger, RL. Scanometric DNA array detection with nanoparticle probes. Science 2000, 289, 1757–1760. [Google Scholar]
- Alivisatos, AP; Johnsson, KP; Peng, X; Wilson, TE; Loweth, CJ; Bruchez, MP, Jr; Schultz, PG. Organization of “nanocrystal molecules” using DNA. Nature 1996, 382, 609–611. [Google Scholar]
- Susuki, K; Hosokawa, K; Maeda, M. Controlling the number and positions of oligonucleotides on gold nanoparticle surfaces. J. Am. Chem. Soc 2009, 131, 7518–7519. [Google Scholar]
- Hill, HD; Hurst, SJ; Mirkin, CA. Curvature-induced base pair “slipping” effects in DNA-nanoparticle hybridization. Nano Lett 2009, 9, 317–321. [Google Scholar]
- Park, SY; Lytton-Jean, AK; Lee, B; Weigand, S; Schatz, GC; Mirkin, CA. DNA-programmable nanoparticle crystallization. Nature 2008, 451, 553–556. [Google Scholar]
- Lee, JS; Ulmann, PA; Han, MS; Mirkin, CA. A DNA-gold nanoparticle-based colorimetric competition assay for the detection of cysteine. Nano Lett 2008, 8, 529–533. [Google Scholar]
- Claridge, SA; Liang, HW; Basu, SR; Frechet, JM; Alivisatos, AP. Isolation of discrete nanoparticle-DNA conjugates for plasmonic applications. Nano Lett 2008, 8, 1202–1206. [Google Scholar]
- Ramos, R; Manning, B; Aviñó, A; Gargallo, R; Eritja, R. Photocleavage of peptides and oligodeoxynucleotides carrying 2-nitrobenzyl groups. Helv. Chim. Acta 2009, 92, 613–622. [Google Scholar]
- Manning, B; Leigh, SJ; Ramos, R; Preece, J; Eritja, R. Fabrication of patterned surfaces by photolithographic exposure of DNA-hairpins carrying a novel photolabile group. J. Exp. Nanosci 2009, 5, 26–39. [Google Scholar]
- Hegner, M; Wagner, P; Semenza, G. Ultralarge atomically flat template-stripped Au surfaces for scanning probe microscopy. Surf. Sci 1993, 291, 39–46. [Google Scholar]
- Mourougou-Candoni, N; Naud, C; Thibaudau, F. Adsorption of thiolated oligonucleotides on gold surfaces: An atomic force microscopy study. Langmuir 2003, 19, 682–686. [Google Scholar]
- Herne, TM; Tarlov, MJ. Characterization of DNA probes immobilized on gold surfaces. J. Am. Chem. Soc 1997, 119, 8916–8920. [Google Scholar]
- Csáki, A; Möller, R; Straube, W; Köhler, JM; Fritzsche, W. DNA monolayer on gold substrates characterized by nanoparticle labeling and scanning force microscopy. Nucleic Acids Res 2001, 29, e81. [Google Scholar]
- Niemeyer, CM; Ceyhan, B; Gao, S; Chi, L; Peschel, S; Simon, U. Site-selective immobilization of gold nanoparticles functionalized with DNA oligomers. Colloid Polym. Sci 2001, 279, 68–72. [Google Scholar]
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Manning, B.; Eritja, R. Use of Oligonucleotides Carrying Photolabile Groups for the Control of the Deposition of Nanoparticles in Surfaces and Nanoparticle Association. Int. J. Mol. Sci. 2011, 12, 7238-7249. https://doi.org/10.3390/ijms12107238
Manning B, Eritja R. Use of Oligonucleotides Carrying Photolabile Groups for the Control of the Deposition of Nanoparticles in Surfaces and Nanoparticle Association. International Journal of Molecular Sciences. 2011; 12(10):7238-7249. https://doi.org/10.3390/ijms12107238
Chicago/Turabian StyleManning, Brendan, and Ramon Eritja. 2011. "Use of Oligonucleotides Carrying Photolabile Groups for the Control of the Deposition of Nanoparticles in Surfaces and Nanoparticle Association" International Journal of Molecular Sciences 12, no. 10: 7238-7249. https://doi.org/10.3390/ijms12107238