Profile and Functional Properties of Seed Proteins from Six Pea (Pisum sativum) Genotypes
Abstract
:1. Introduction
2. Experimental Section
2.1. Material
2.2. Protein Content Determination
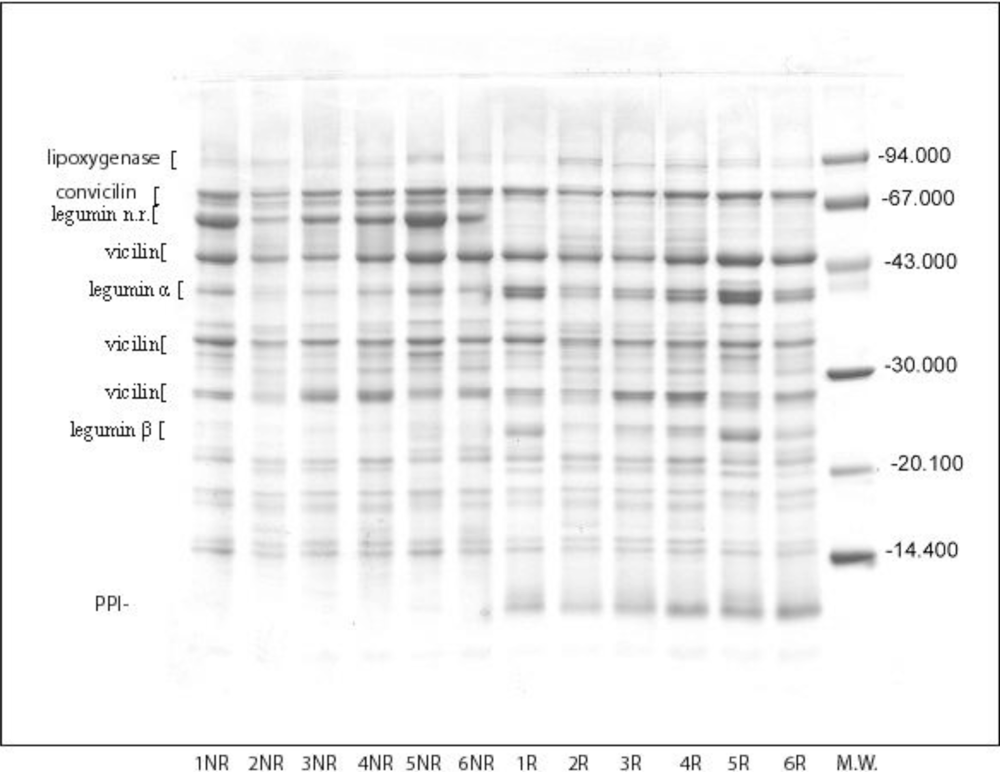
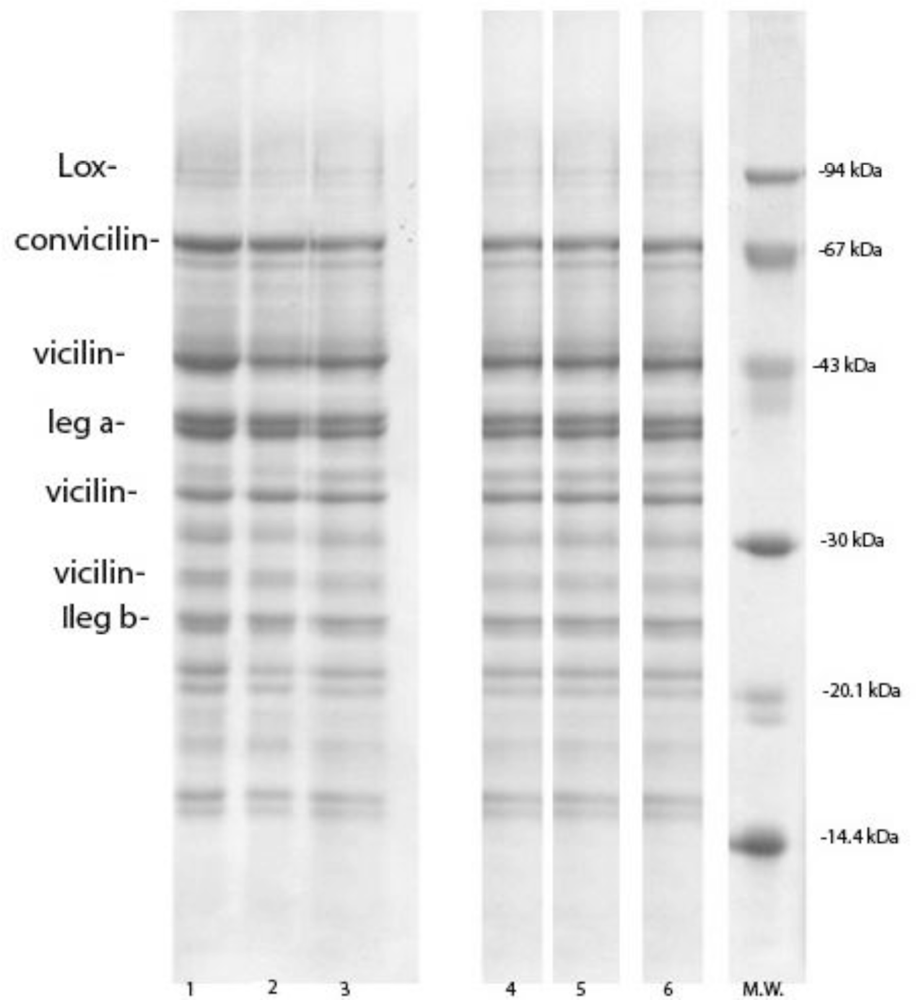
2.3. SDS-PAGE
2.4. Densitometric Analysis
2.5. Protein Solubility
2.6. Emulsifying Properties
2.7. Foaming Properties
2.8. Statistical Analysis
3. Results and Discussion
3.1. Extractibility of Pea Bean Proteins
3.2. Pea Bean Protein Composition
3.3. Pea Protein Isolates Composition
3.4. Solubility
3.5. Emulsifying Properties
3.6. Foaming Properties
4. Conclusions
Acknowledgments
References
- Roy, F; Boye, JI; Simpson, BK. Bioactive proteins and peptides in pulse crops: Pea, chickpea and lentil. Food Res. Int 2010, 43, 432–442. [Google Scholar]
- Gueguen, JCM; Barbot, J; Schaeffer, F. Dissociation and aggregation of pea legumin induced by pH and ionic strength. J. Sci. Food Agric 1988, 53, 167–182. [Google Scholar]
- Heng, L; van Koningsveld, GA; Gruppen, H; van Boekel, MAJS; Vincken, JP; Roozen, JP; Voragen, AGJ. Protein-flavour interactions in relation to development of novel protein foods. Trends Food Sci. Technol 2004, 15, 217–224. [Google Scholar]
- Shewry, PR; Napier, JA; Tatham, AS. Seed Storage Proteins: Structures and BiosynthESIs. Plant Cell 1995, 7, 945–956. [Google Scholar]
- Griga, M; Horáček, J; Klenotičová, H. Protein patterns associated with Pisum sativum somatic embryogenESIs. Biol. Plantarum 2007, 51, 201–211. [Google Scholar]
- Croy, RR; Gatehouse, JA; Tyler, M; Boulter, D. The purification and characterization of a third storage protein (convicilin) from the seeds of pea (Pisum sativum L.). Biochem. J 1980, 191, 509–516. [Google Scholar]
- O’Kane, FE; Happe, RP; Vereijken, JM; Gruppen, H; van Boekel, MAJS. Characterization of Pea Vicilin. 1. Denoting Convicilin as the α-Subunit of the Pisum Vicilin Family. J. Agric. Food Chem 2004, 52, 3141–3148. [Google Scholar]
- Schroeder, HE. Quantitative studies on the cotyledonary proteins in the genus Pisum. J. Agric. Food Chem 1982, 33, 623–633. [Google Scholar]
- Bora, SP; Breeke, JC; Powers, RJ. Heat Induced Gelation of Peas (Pisum sativum) Mixed Globulins, Vicilin and Legumin; Institute of Food Technologists: Chicago, IL, USA, 1994; Volume 59. [Google Scholar]
- O’Kane, FE; Happe, RP; Vereijken, JM; Gruppen, H; van Boekel, MAJS. Heat-Induced Gelation of Pea Legumin: Comparison with Soybean Glycinin. J. Agric. Food Chem 2004, 52, 5071–5078. [Google Scholar]
- O’Kane, FE; Vereijken, JM; Gruppen, H; van Boekel, MAJS. Gelation behavior of protein isolates extracted from 5 cultivars of Pisum sativum L. J. Food Sci 2005, 70, C132–C137. [Google Scholar]
- Cserhalmi, Z; Czukor, B; Gajzágó-Schuster, I. Emulsifying properties, surface hydrophobicity and thermal denaturation of pea protein fractions. Acta Aliment 1998, 27, 357–363. [Google Scholar]
- Rangel, A; Domont, GB; Pedrosa, C; Ferreira, ST. Functional Properties of Purified Vicilins from Cowpea (Vigna unguiculata) and Pea (Pisum sativum) and Cowpea Protein Isolate. J. Agric. Food Chem 2003, 51, 5792–5797. [Google Scholar]
- Kimura, A; Fukuda, T; Zhang, M; Motoyama, S; Maruyama, N; Utsumi, S. Comparison of Physicochemical Properties of 7S and 11S Globulins from Pea, Fava Bean, Cowpea, and French Bean with Those of Soybean French Bean 7S Globulin Exhibits Excellent Properties. J. Agric. Food Chem 2008, 56, 10273–10279. [Google Scholar]
- Martínez-Villaluenga, C; Gulewicz, P; Frias, J; Gulewicz, K; Vidal-Valverde, C. Assessment of protein fractions of three cultivars of Pisum sativum L.: Effect of germination. Eur. Food Res. Technol 2008, 226, 1465–1478. [Google Scholar]
- Guleria, S; Dua, S; Chongtham, N. Analysis of variability in different genotypes of pea (Pisum sativum L.) on the basis of protein markers. Legume Res 2009, 32, 265–269. [Google Scholar]
- Wang, N; Hatcher, DW; Warkentin, TD; Toews, R. Effect of cultivar and environment on physicochemical and cooking characteristics of field pea (Pisum sativum). Food Chem 2010, 118, 109–115. [Google Scholar]
- Nikolopoulou, D; Grigorakis, K; Stasini, M; Alexis, MN; Iliadis, K. Differences in chemical composition of field pea (Pisum sativum) cultivars: Effects of cultivation area and year. Food Chem 2007, 103, 847–852. [Google Scholar]
- Maninder, K; Sandhu, KS; Singh, N. Comparative study of the functional, thermal and pasting properties of flours from different field pea (Pisum sativum L.) and pigeon pea (Cajanus cajan L.) cultivars. Food Chem 2007, 104, 259–267. [Google Scholar]
- Riblett, AL; Herald, TJ; Schmidt, KA; Tilley, KA. Characterization of β-Conglycinin and Glycinin Soy Protein Fractions from Four Selected Soybean Genotypes. J. Agric. Food Chem 2001, 49, 4983–4989. [Google Scholar]
- Khatib, KA; Herald, TJ; Aramouni, FM; MacRitchie, F; Schapaugh, WT. Characterization and Functional Properties of Soy β-Conglycinin and Glycinin of Selected Genotypes. J. Food Sci 2002, 67, 2923–2929. [Google Scholar]
- PESIc, MB; Vucelic-Radovic, BV; Barac, MB; Stanojevic, SP. The influence of genotypic variation in protein composition on emulsifying properties of soy proteins. J. Am. Oil Chem. Soc 2005, 82, 667–672. [Google Scholar]
- Periago, MJ; Vidal, ML; Ros, G; Rincón, F; Martínez, C; López, G; Rodrigo, J; Martínez, I. Influence of enzymatic treatment on the nutritional and functional properties of pea flour. Food Chem 1998, 63, 71–78. [Google Scholar]
- Pedrosa, C; Trisciuzzi, C; Ferreira, ST. Effects of Glycosylation on Functional Properties of Vicilin, the 7S Storage Globulin from Pea (Pisum sativum). J. Agric. Food Chem 1997, 45, 2025–2030. [Google Scholar]
- Tömösközi, S; Lásztity, R; Haraszi, R; Baticz, O. Isolation and study of the functional properties of pea proteins. Nahrung 2001, 45, 399–401. [Google Scholar]
- Fuhrmeister, H; Meuser, F. Impact of processing on functional properties of protein products from wrinkled peas. J. Food Eng 2003, 56, 119–129. [Google Scholar]
- Dua, S; Mahajan, A; Sandal, M; Gagan. Physico-chemical properties of defatted pea seed meal proteins with emphasis on salts and pH. J. Food Sci. Tech 2009, 46, 251–254. [Google Scholar]
- Sumner, AK; Nielsen, MA; Youngs, CG. Production and Evaluation of Pea Protein Isolate. J. Food Sci 1981, 46, 364–366. [Google Scholar]
- Vu Huu, T; Shibasaki, K. Major proteins of soybean seeds. A straightforward fractionation and their characterization. J. Agric. Food Chem 1976, 24, 1117–1121. [Google Scholar]
- Bradford, MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem 1976, 72, 248–254. [Google Scholar]
- Fling, SP; Gregerson, DS. Peptide and protein molecular weight determination by electrophorESIs using a high-molarity tris buffer system without urea. Anal. Biochem 1986, 155, 83–88. [Google Scholar]
- Cai, T; Chang, KC. Processing Effect on Soybean Storage Proteins and Their Relationship with Tofu Quality. J. Agric. Food Chem 1999, 47, 720–727. [Google Scholar]
- Wu, W; Hettiarachchy, N; Qi, M. Hydrophobicity, solubility, and emulsifying properties of soy protein peptides prepared by papain modification and ultrafiltration. J. Am. Oil Chem. Soc 1998, 75, 845–850. [Google Scholar]
- Sathe, SK; Deshpande, SS; Salunke, DK. Functional properties of lupin seeds (Lupinus mutabilis) proteins and protein concentrates. J. Food Sci 1981, 9, 491–502. [Google Scholar]
- Alonso, R; Orúe, E; Zabalza, MJ; Grant, G; Marzo, F. Effect of extrusion cooking on structure and functional properties of pea and kidney bean proteins. J. Sci. Food Agric 2000, 80, 397–403. [Google Scholar]
- Szymanowska, U; Jakubczyk, A; Baraniak, B; Kur, A. Characterisation of lipoxygenase from pea seeds (Pisum sativum varTelephone L.). Food Chem 2009, 116, 906–910. [Google Scholar]
- Shand, PJ; Ya, H; Pietrasik, Z; Wanasundara, PKJPD. Physicochemical and textural properties of heat-induced pea protein isolate gels. Food Chem 2007, 102, 1119–1130. [Google Scholar]
- Barac, M; Stanojevic, S. The effect of microwave roasting on soybean protein composition and components with trypsin inhibitor activity. Acta Aliment 2005, 34, 23–31. [Google Scholar]
- Aluko, RE; McIntosh, T. Polypeptide profile and functional properties of defatted meals and protein isolates of canola seeds. J. Sci. Food Agric 2001, 81, 391–396. [Google Scholar]
- Aluko, RE; Mofolasayo, OA; Watts, BM. Emulsifying and Foaming Properties of Commercial Yellow Pea (Pisum sativum L.) Seed Flours. J. Agric. Food Chem 2009, 57, 9793–9800. [Google Scholar]
- Tsoukala, A; Papalamprou, E; Makri, E; Doxastakis, G; Braudo, EE. Adsorption at the air-water interface and emulsification properties of grain legume protein derivatives from pea and broad bean. Colloids Surf. B 2006, 53, 203–208. [Google Scholar]
- Belitz, HD; Grosch, W; Schieberle, P. Food Chemistry; Springer-verlag: Berlin, Germany, 2009; p. 989. [Google Scholar]
- Boye, JI; Aksay, S; Roufik, S; Ribéreau, S; Mondor, M; Farnworth, E; Rajamohamed, SH. Comparison of the functional properties of pea, chickpea and lentil protein concentrates processed using ultrafiltration and isoelectric precipitation techniques. Food Res. Int 2010, 43, 537–546. [Google Scholar]




| Varieties | Total Proteina Pea Bean (g/kg) | Extractable Protein Contenta (g/kg) | Extractabilityb | Total Proteina Isolate (g/kg) | ||
|---|---|---|---|---|---|---|
| Tris-HCl pH 8.0 | Water pH 8.0 | Tris-HCl pH 8.0 | Water pH 8.0 | |||
| Calvedon | 244.21 ± 0.21a | 147.81 ± 1.2a | 107.42 ± 0.20a | 605.2 ± 4.9a | 439.8 ± 0.8a | 837.71 ± 2.13a |
| L1 | 317.63 ± 0.29b | 227.11 ± 0.62b | 117.20 ± 1.01b | 714.7 ± 1.9b | 369.0 ± 3.1b | 846.65 ± 1.54b |
| L2 | 241.42 ± 0.11c | 142.20 ± 0.21c | 97.61 ± 0.41c | 589.0 ± 0.8c | 404.3 ± 1.3c | 835.09 ± 0.72a |
| L3 | 233.40 ± 0.3d | 150.63 ± 0.82a | 92.01 ± 0.6d | 645.2 ± 3.4d | 394.2 ± 2.1d,g | 842.22 ± 0.67a |
| Maja | 273.70 ± 0.10e | 148.65 ± 1.43a | 115.41 ± 0.8b | 542.9 ± 3.4e | 421.6 ± 2.8e | 890.26 ± 2.47b |
| M.A. | 223.31f | 116,64 ± 0.22e | 87.4 ± 0.4e | 522.1 ± 0.9f | 391.4 ± 1.8f,g | 841.53 ± 1.09a |
| Protein | M.w.(kDa) | Concentration (g/kg) | |||||
|---|---|---|---|---|---|---|---|
| Calvedon | L1 | L2 | L3 | Maja | Miracle of America | ||
| Reduced Conditions | |||||||
| Lipoxygenase | 92.7 | 25.5 ± 0.8a | 48.1 ± 1.4b | 25.9 ± 1.0c,a | 29.4 ± 1.3d,b | 24.5 ± 1.7e,a,c | 17.4 ± 0.5f |
| Convicilin | 77.9−72.4 | 83.8a | 71.6 ± 2.0b | 79.7 ± 0.6c | 80.8 ± 1.7d,c | 83.3 ± 1.4a,d | 89.8 ± 2.2e |
| Vicilin | 47.3 | 106.7 ± 1.1a | 112.0 ± 1.0b | 96.7 ± 2.0c | 107.5 ± 0.2d,a | 117.6 ± 0.7e | 126.3 ± 0.3f |
| 37−31.8 | 159.6 ± 0.4a | 181.9b | 144.7 ± 1.7c | 165.0 ± 0.7d | 171.8 ± 0.4e | 151.7 ± 1.2f,c | |
| 28.7 | 75.6 ± 0.7a | 83.4 ± 1.4b | 103.5 ± 0.9c | 102.6 ± 1.3d,c | 79.7 ± 2.4e,a | 89.3 ± 1.7f,b | |
| Σ vicilin | 341.9 ± 1.7a | 377.3 ± 1.3b | 344.9 ± 1.6c,a | 375.1 ± 1.8d,b | 369.1 ± 1.5e | 367.8 ± 2.1f,e | |
| Legumin α | 40.89 | 122.1 ± 0.4a | 80.4 ± 1.7b | 86.3 ± 1.2c | 98.4 ± 0.1d | 151.2 ± 0.6e | 102.8 ± 2.0d |
| Legumin β | 23.1−22.3 | 129.7 ± 0.3a | 96.4 ± 1.5b | 125.8 ± 2.1c,a | 127.2 ± 1.9d,a,c | 157.6 ± 2.7e | 166.1 ± 1.6f |
| Leguminn.r | 63.6 | 33.6 ± 1.1a | 75.6 ± 0.3b | 51.6 ± 0.9c | 55.5d | 38.9 ± 1.0e | 36.6 ± 0.2f,e,a |
| Σ legumin | 285.4 ± 1.9a | 252.4 ± 2.0b | 264.7 ± 3.2c | 281.1 ± 2.7a | 347.7 ± 2.6d | 305.5 ± 1.9e | |
| V/L** | 1.20 ± 0.0012a | 1.49 ± 0.007b | 1.30 ± 0.01c | 1.33 ± 0.006d | 1.06 ± 0.0036e | 1.20 ± 0.0006f | |
| V + C/L** | 1.49 ± 0.0017a | 1.78 ± 0.0017b | 1.60 ± 0.014c | 1.62 ± 0.003d | 1.30 ± 0.0027e | 1.50 ± 0.002a | |
| Legn.r/Legr | 0.13a | 0.42b | 0.24c | 0.25c | 0.13e,a | 0.14a,e | |
| PPI | 11.5 | 7.73 ± 0.23a | 6.67 ± 0.11b | 7.55 ± 0.15c,a | 7.05 ± 0.25d | 8.02 ± 0.09e | 10.81 ± 0.27f |
| Non Reduced Conditions | |||||||
| lypoxigenase | 92.7 | 31.3 ± 0.2a | 43.3 ± 1.1b | 35.3 ± 0.7c | 31.5 ± 0.4a | 40.1 ± 0.1e | 27.3 ± 0.6f |
| PPI*** | 11.5 | 9.4 ± 0.1a | 1.0b | 9.0 ± 0.1a | 6.2 ± 0.2d | 3.6e | 2.1 ± 0.3f,b,e |
| convicilin | 77.9−72.4 | 112.0 ± 2.1a | 112.5 ± 1.0a | 118.5 ± 1.1b | 114.4 ± 2.0a,b | 123.4 ± 2.6c | 215.1 ± 4.2d |
| vicilin | 47.3 | 128.8 ± 0.9a | 150.5 ± 2.3b | 103.9 ± 0.7c | 111.5 ± 1.5d | 122.0 ± 1.1e | 57.2 ± 1.4f |
| 37−31.8 | 189.1 ± 1.9a | 178.2 ± 2.2b | 170.8 ± 1.2c | 188.5 ± 0.6d,a | 197.6 ± 0.8e | 220.2 ± 0.4f | |
| 28.7 | 61.1 ± 0.2a | 66.5 ± 1.0b | 110.5 ± 0.3c | 109.0 ± 1.3c | 68.3 ± 2.4e,b | 22.2 ± 0.9f | |
| Σ vicilin | 379.0 ± 3.0a | 395.2 ± 5.5b | 385.2 ± 2.2c | 409.0 ± 3.4d | 387.9 ± 4.3b,c | 299.6 ± 2.7f | |
| Legumin α | 40.89 | 65.3 ± 1.0a | 63.2 ± 2.7b,a | 44.4 ± 0.1c | 53.6 ± 2.2d | 74.4 ± 1.2e | 42.4 ± 1.1f,c |
| Legumin β | 23.1−22.3 | 72.8 ± 0.8a | 58.0 ± 1.4b | 59.0 ± 1.9c,b | 56.8 ± 0.3d,b,c | 73.0 ± 2.0e | 61.8 ± 0.7f,c |
| Legn.r. | 63.6 | 152.7 ± 2.5a | 75.4 ± 1.1b | 122.5 ± 1.6c | 106.6 ± 1.5d | 155.8 ± 0.9e,a | 127.5 ± 1.5f,c |
| Σ Leg | 290.8 ± 4.3a | 196.6 ± 5.1b | 225.9 ± 3.6c | 217.0 ± 4.0d | 303.2 ± 4.1e | 231.7 ± 2.6c | |
| Legumin α | 40.89 | 65.3 ± 1.0a | 63.2 ± 2.7b,a | 44.4 ± 0.1c | 53.6 ± 2.2d | 74.4 ± 1.2e | 42.4 ± 1.1f,c |
| V/L | 1.30 ± 0.009a | 2.09 ± 0.026b | 1.70 ± 0.017c | 1.88 ± 0.019d | 1.28 ±0.022e | 1.29 ± 0.0027a | |
| C + V/L | 1.69 ± 0.031a | 2.54 ± 0.036b | 2.23 ± 0.021c | 2.41 ± 0.019d | 1.69 ± 0.021a | 2.22 ± 0.005c | |
| Legn.r/Legr | 1.11 ± 0.03a | 0.62b | 1.18 ± 0,02a | 0.97 ± 0.04d | 1.06 ± 0.07a | 1.22c | |
| Protein/polypeptide | M.w.(kDa) | Concentration (g/kg) | |||||
|---|---|---|---|---|---|---|---|
| Calvedon | L1 | L2 | L3 | Maja | Miracle of America | ||
| Isolate | |||||||
| lipoxygenase | 92.7 | 29.8 ± 0.7a | 25.2 ± 0.2b | 34.3 ± 0.4c | 43.5 ± 1.0d | 36.8 ± 1.4c | 27.7 ± 0.5a |
| convicilin | 77.9−72.4 | 104.2 ± 0.1a | 102.7 ± 1.1a | 117.8 ± 2.6b | 103.8 ± 1.6a | 91.3 ± 0.6c | 97.4 ± 1.2d |
| Vicilin | 47.3 | 153.1 ± 4.3a | 171.8 ± 0.9b | 168.3 ± 2.7b,c | 176.8 ± 3.4d | 123.0 ± 1.1e | 166.7 ± 2.4b |
| Vicilin | 37−31.8 | 195.6 ± 1.7a | 207.0 ± 3.3b | 194.1 ± 3.1a | 207.7 ± 2.2b | 177.5 ± 3.6c | 188.7 ± 1.8d |
| Σ vicilin | 348.7 ± 6.0e | 378.8 ± 4.2a | 362.4 ± 5.8b | 384.5 ± 5.0c | 300.5 ± 4.7d | 355.4 ± 4.2f,b | |
| legumin α | 40.89 | 133.5 ± 2.6a | 162.3 ± 2.1b | 155.6 ± 3.9c | 95.6 ± 2.9d | 115.3 ± 1.1e | 142.0 ± 0.7f |
| Legumin β | 23.1−22.3 | 144.2 ± 0.2a | 142.7 ± 0.7b | 142.0 ± 0.4b | 152.8 ± 2.2c | 168.2 ± 1.7d | 147.0 ± 0.3,e |
| Legn.r | 63.6 | 35.4 ± 0.7a | 24.6 ± 0.2b | 36.8 ± 0.5a | 30.8 ± 0.4c | 34.3 ± 2.1a | 32.7 ± 0.4f,c,d,e |
| Σ legumin | 313.1 ± 3.5a | 329.6 ± 3.3b | 334.4 ± 5.0b | 280.2 ± 4.9c | 317.8 ± 4.9,da | 321.7 ± 1.4d | |
| V/L | 1.11 ± 0.0067a | 1.15 ± 0.0018b | 1.08 ± 0.0011c | 1.37 ± 0.0061c | 0.94 ± 0.0002d | 1.10 ± 0.0082c | |
| C+V/L | 1.45 ± 0.003a | 1.46 ± 0.002b | 1.43 ± 0.0036c | 1.74 ± 0.0069d | 1.23 ± 0.0023e | 1.41 ± 0.01066f | |
| PPI | 17.9 ± 0.8a | 17.0 ± 0.2a | 26.2 ± 0.5c | 17.6 ± 0.5a | 14.0 ± 0.7d | 21.0f | |
| whey | |||||||
| lipoxygenase | 92.7 | 33.7 ± 0.9a | 22.2 ± 0.4b | 26.2 ± 0.2c | 29.8d | 64.4 ± 0.8e | 32.4 ± 0.8a |
| Legumin α | 40.89 | 45.20 ± 0.4a | 58.9 ± 0.3b | 50.1 ± 0.1c | 54.9 ± 0.7d | 38.1 ± 0.2e | 47.1 ± 0.4f |
| vicilin | 28.5 | 154.2 ± 1.1a | 131.0 ± 1.0b | 168.7 ± 1.2c | 173.3 ± 2.5d | 123.7 ± 2.4e | 140.2 ± 2.1f |
| PPI | 11.5 | 261.1 ± 1.7a | 349.1 ± 1.4b | 242.0 ± 2.20c | 324.6 ± 2.7d | 229.2 ± 2.7e | 253.3 ± 3.7f |
© 2010 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Barac, M.; Cabrilo, S.; Pesic, M.; Stanojevic, S.; Zilic, S.; Macej, O.; Ristic, N. Profile and Functional Properties of Seed Proteins from Six Pea (Pisum sativum) Genotypes. Int. J. Mol. Sci. 2010, 11, 4973-4990. https://doi.org/10.3390/ijms11124973
Barac M, Cabrilo S, Pesic M, Stanojevic S, Zilic S, Macej O, Ristic N. Profile and Functional Properties of Seed Proteins from Six Pea (Pisum sativum) Genotypes. International Journal of Molecular Sciences. 2010; 11(12):4973-4990. https://doi.org/10.3390/ijms11124973
Chicago/Turabian StyleBarac, Miroljub, Slavica Cabrilo, Mirjana Pesic, Sladjana Stanojevic, Sladjana Zilic, Ognjen Macej, and Nikola Ristic. 2010. "Profile and Functional Properties of Seed Proteins from Six Pea (Pisum sativum) Genotypes" International Journal of Molecular Sciences 11, no. 12: 4973-4990. https://doi.org/10.3390/ijms11124973
APA StyleBarac, M., Cabrilo, S., Pesic, M., Stanojevic, S., Zilic, S., Macej, O., & Ristic, N. (2010). Profile and Functional Properties of Seed Proteins from Six Pea (Pisum sativum) Genotypes. International Journal of Molecular Sciences, 11(12), 4973-4990. https://doi.org/10.3390/ijms11124973






