In-Silico Analysis of Phytocompounds of Olea europaea as Potential Anti-Cancer Agents to Target PKM2 Protein
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of the Phytocompounds Database
2.2. Prediction of ADMET Properties
2.3. Prediction of Anticancer Activity
2.4. Retrieval and Preparation of Protein
2.5. Screening of Potential Lead Compounds
2.6. Molecular Dynamics Simulations
3. Results and Discussion
3.1. Prediction of ADMET Properties
3.2. Prediction of Anticancer Activity
3.3. Screening of Potential Phytocompounds
3.4. Molecular Dynamics Simulation
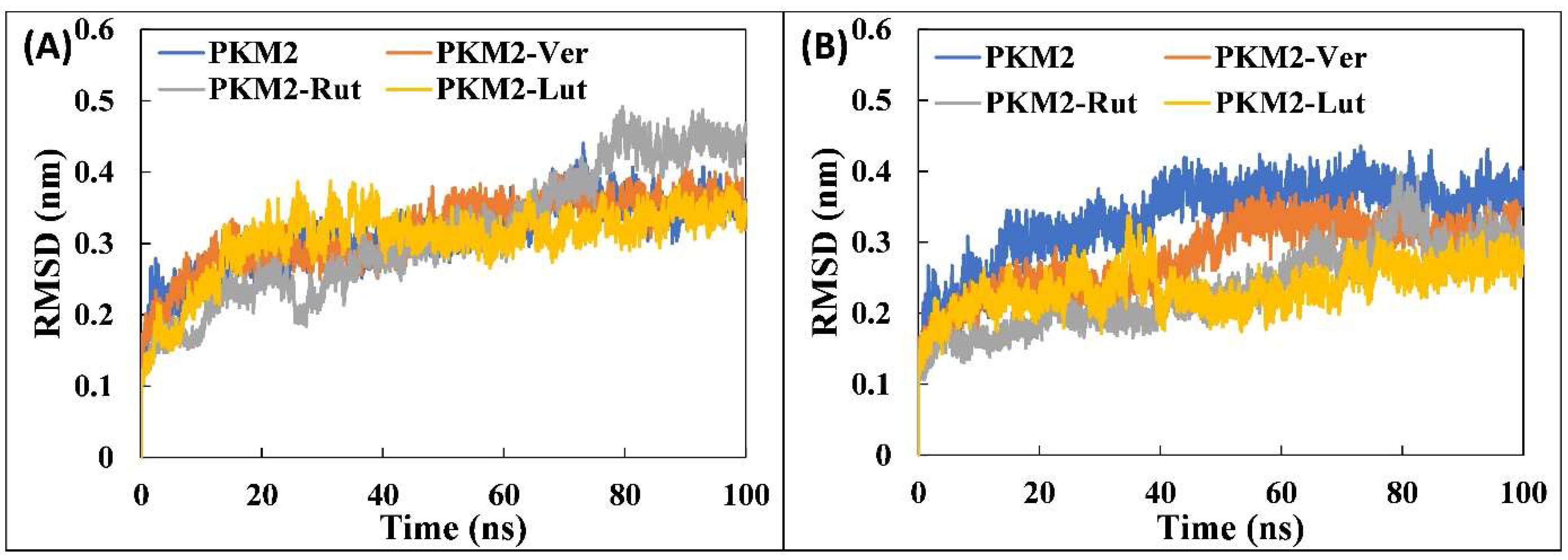
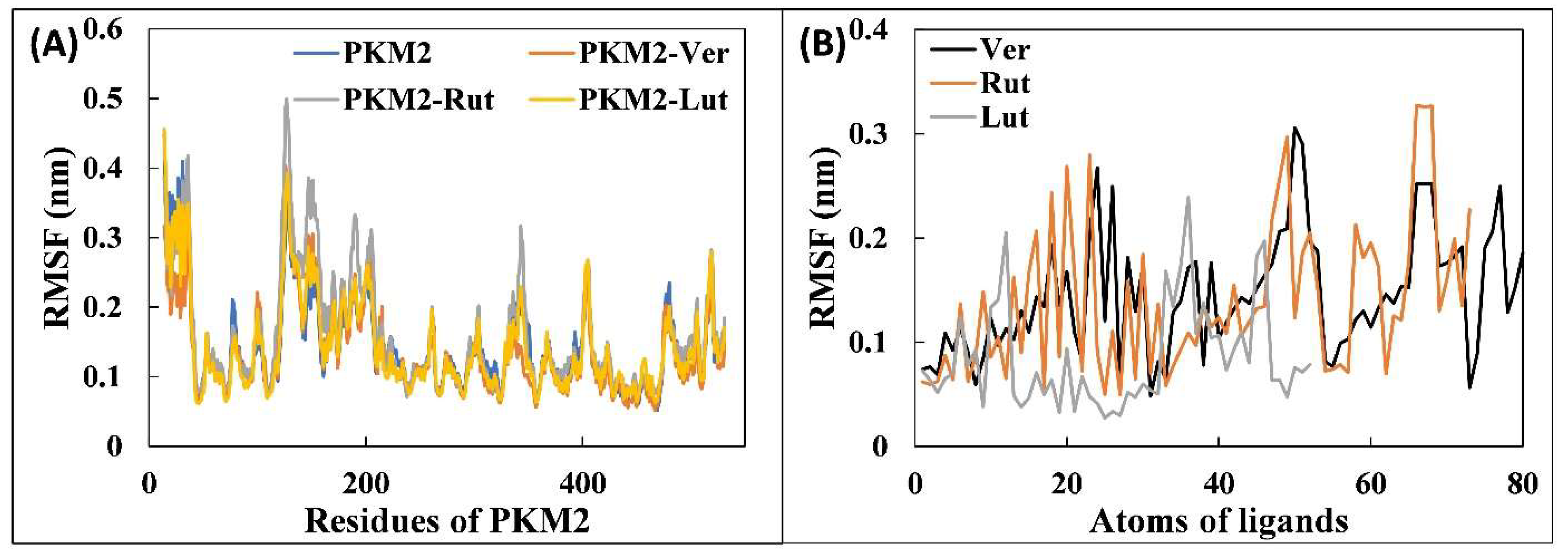
3.4.1. Analysis of RMSD and RMSF
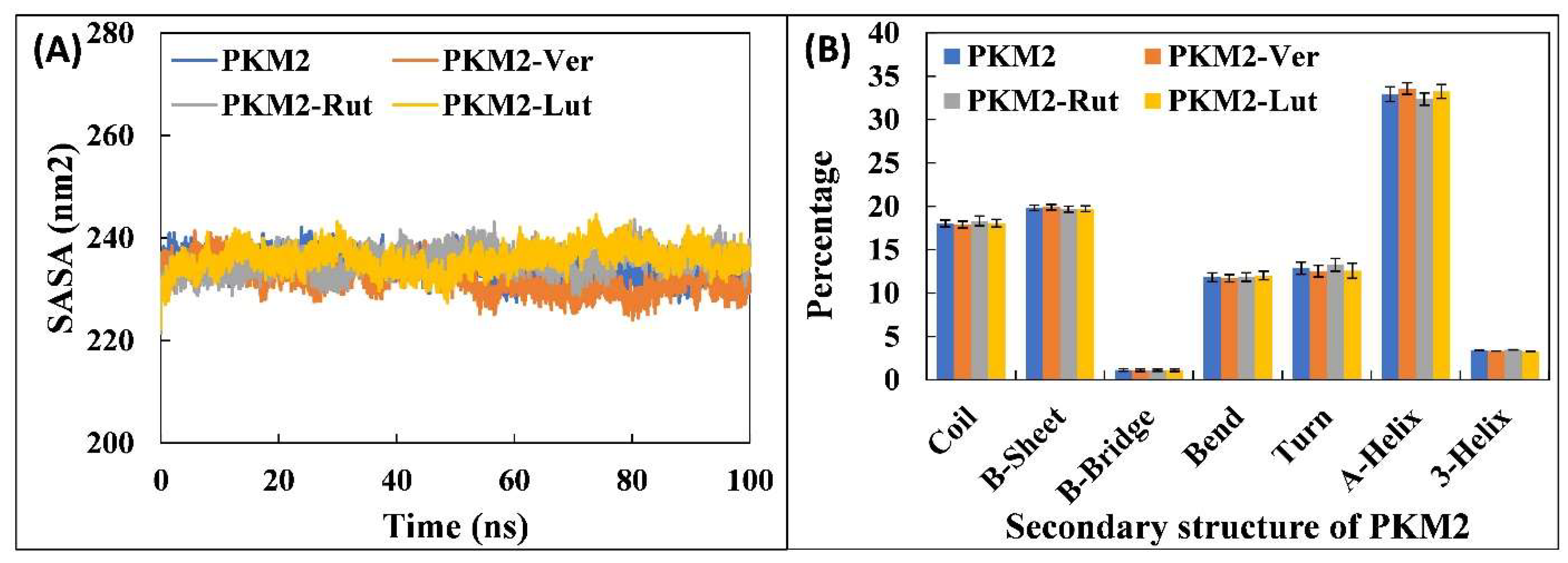
3.4.2. Analysis of Physicochemical Parameters, Structure Compactness and Secondary Structure
3.4.3. Analysis of Hydrogen Bonds and Binding Energies
3.5. Principal Component Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
Abbreviations
| ADME | Absorption, distribution, metabolism, excretion |
| Lut | Luteolin_7_O_glucoside |
| OLE | Olive leaves extract |
| PKM2 | Pyruvate kinase M2 isoform |
| Rut | Rutin |
| Ver | Verbascoside |
References
- Globocan GLOBOCAN 2020: New Global Cancer Data. Available online: https://www.uicc.org/news/globocan-2020-new-global-cancer-data (accessed on 26 August 2022).
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Liberti, M.V.; Locasale, J.W. The Warburg Effect: How Does It Benefit Cancer Cells? Trends Biochem. Sci. 2016, 41, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Guo, Y.; Zhang, X.; Liu, H.; Yin, M.; Chen, X.; Peng, C. Pyruvate Kinase M2 (PKM2) in Cancer and Cancer Therapeutics. Cancer Lett. 2021, 503, 240–248. [Google Scholar] [CrossRef]
- Dayton, T.L.; Jacks, T.; Vander Heiden, M.G. PKM 2, Cancer Metabolism, and the Road Ahead. EMBO Rep. 2016, 17, 1721–1730. [Google Scholar] [CrossRef] [PubMed]
- Keller, K.E.; Tan, I.S.; Lee, Y.-S. SAICAR Stimulates Pyruvate Kinase Isoform M2 and Promotes Cancer Cell Survival in Glucose-Limited Conditions. Science 2012, 338, 1069–1072. [Google Scholar] [CrossRef] [PubMed]
- Talhaoui, N.; Taamalli, A.; Gómez-Caravaca, A.M.; Fernández-Gutiérrez, A.; Segura-Carretero, A. Phenolic Compounds in Olive Leaves: Analytical Determination, Biotic and Abiotic Influence, and Health Benefits. Food Res. Int. 2015, 77, 92–108. [Google Scholar] [CrossRef]
- Mihailova, A.; Abbado, D.; Pedentchouk, N. Differences in n -Alkane Profiles between Olives and Olive Leaves as Potential Indicators for the Assessment of Olive Leaf Presence in Virgin Olive Oils. Eur. J. Lipid Sci. Technol. 2015, 117, 1480–1485. [Google Scholar] [CrossRef]
- Boss, A.; Bishop, K.; Marlow, G.; Barnett, M.; Ferguson, L. Evidence to Support the Anti-Cancer Effect of Olive Leaf Extract and Future Directions. Nutrients 2016, 8, 513. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Qu, J.; Luo, S.; Feng, S.; Li, T.; Yuan, M.; Huang, Y.; Liao, J.; Yang, R.; Ding, C. Optimization of Ultrasound-Assisted Extraction of Flavonoids from Olive (Olea europaea) Leaves, and Evaluation of Their Antioxidant and Anticancer Activities. Molecules 2018, 23, 2513. [Google Scholar] [CrossRef]
- Salama, Z.A.; Aboul-Enein, A.M.; Gaafar, A.A.; Asker, M.S.; Aly, H.F.; Ahmed, H.A. In-Vitro Antioxidant, Antimicrobial and Anticancer Activities of Banana Leaves ( Musa Acuminata ) and Olive Leaves (Olea europaea L.) as by-Products. Res. J. Pharm. Technol. 2020, 13, 687. [Google Scholar] [CrossRef]
- Antoniou, C.; Hull, J. The Anti-Cancer Effect of Olea europaea L. Products: A Review. Curr. Nutr. Rep. 2021, 10, 99–124. [Google Scholar] [CrossRef]
- Fu, S.; Arráez-Roman, D.; Segura-Carretero, A.; Menéndez, J.A.; Menéndez-Gutiérrez, M.P.; Micol, V.; Fernández-Gutiérrez, A. Qualitative Screening of Phenolic Compounds in Olive Leaf Extracts by Hyphenated Liquid Chromatography and Preliminary Evaluation of Cytotoxic Activity against Human Breast Cancer Cells. Anal. Bioanal. Chem. 2010, 397, 643–654. [Google Scholar] [CrossRef]
- Quirantes-Piné, R.; Lozano-Sánchez, J.; Herrero, M.; Ibáñez, E.; Segura-Carretero, A.; Fernández-Gutiérrez, A. HPLC-ESI-QTOF-MS as a Powerful Analytical Tool for Characterising Phenolic Compounds in Olive-Leaf Extracts. Phytochem. Anal. 2013, 24, 213–223. [Google Scholar] [CrossRef]
- Martín-García, B.; Verardo, V.; León, L.; De la Rosa, R.; Arráez-Román, D.; Segura-Carretero, A.; Gómez-Caravaca, A.M. GC-QTOF-MS as Valuable Tool to Evaluate the Influence of Cultivar and Sample Time on Olive Leaves Triterpenic Components. Food Res. Int. 2019, 115, 219–226. [Google Scholar] [CrossRef]
- Olmo-García, L.; Kessler, N.; Neuweger, H.; Wendt, K.; Olmo-Peinado, J.; Fernández-Gutiérrez, A.; Baessmann, C.; Carrasco-Pancorbo, A. Unravelling the Distribution of Secondary Metabolites in Olea europaea L.: Exhaustive Characterization of Eight Olive-Tree Derived Matrices by Complementary Platforms (LC-ESI/APCI-MS and GC-APCI-MS). Molecules 2018, 23, 2419. [Google Scholar] [CrossRef] [PubMed]
- Alañón, M.E.; Ivanović, M.; Gómez-Caravaca, A.M.; Arráez-Román, D.; Segura-Carretero, A. Choline Chloride Derivative-Based Deep Eutectic Liquids as Novel Green Alternative Solvents for Extraction of Phenolic Compounds from Olive Leaf. Arab. J. Chem. 2020, 13, 1685–1701. [Google Scholar] [CrossRef]
- Meirinhos, J.; Silva, B.M.; ValentÃo, P.; Seabra, R.M.; Pereira, J.A.; Dias, A.; Andrade, P.B.; Ferreres, F. Analysis and Quantification of Flavonoidic Compounds from Portuguese Olive (Olea europaea L.) Leaf Cultivars. Nat. Prod. Res. 2005, 19, 189–195. [Google Scholar] [CrossRef]
- Ghomari, O.; Sounni, F.; Massaoudi, Y.; Ghanam, J.; Drissi Kaitouni, L.B.; Merzouki, M.; Benlemlih, M. Phenolic Profile (HPLC-UV) of Olive Leaves According to Extraction Procedure and Assessment of Antibacterial Activity. Biotechnol. Rep. 2019, 23, e00347. [Google Scholar] [CrossRef]
- Brahmi, F.; Mechri, B.; Dhibi, M.; Hammami, M. Variations in Phenolic Compounds and Antiradical Scavenging Activity of Olea europaea Leaves and Fruits Extracts Collected in Two Different Seasons. Ind. Crops Prod. 2013, 49, 256–264. [Google Scholar] [CrossRef]
- Servili, M.; Sordini, B.; Esposto, S.; Urbani, S.; Veneziani, G.; Di Maio, I.; Selvaggini, R.; Taticchi, A. Biological Activities of Phenolic Compounds of Extra Virgin Olive Oil. Antioxidants 2013, 3, 1–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ortega-García, F.; Peragón, J. Phenol Metabolism in the Leaves of the Olive Tree (Olea europaea L.) Cv. Picual, Verdial, Arbequina, and Frantoio during Ripening. J. Agric. Food Chem. 2010, 58, 12440–12448. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera?A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Pires, D.E.V.; Blundell, T.L.; Ascher, D.B. PkCSM: Predicting Small-Molecule Pharmacokinetic and Toxicity Properties Using Graph-Based Signatures. J. Med. Chem. 2015, 58, 4066–4072. [Google Scholar] [CrossRef]
- Wang, Y.; Xing, J.; Xu, Y.; Zhou, N.; Peng, J.; Xiong, Z.; Liu, X.; Luo, X.; Luo, C.; Chen, K.; et al. In Silico ADME/T Modelling for Rational Drug Design. Q. Rev. Biophys. 2015, 48, 488–515. [Google Scholar] [CrossRef]
- Filimonov, D.A.; Lagunin, A.A.; Gloriozova, T.A.; Rudik, A.V.; Druzhilovskii, D.S.; Pogodin, P.V.; Poroikov, V.V. Prediction of the Biological Activity Spectra of Organic Compounds Using the Pass Online Web Resource. Chem. Heterocycl. Compd. 2014, 50, 444–457. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2009, 31, 455–461. [Google Scholar] [CrossRef]
- Wallace, A.C.; Laskowski, R.A.; Thornton, J.M. LIGPLOT: A Program to Generate Schematic Diagrams of Protein-Ligand Interactions The LIGPLOT Program Automatically Generates Schematic 2-D Representations of Protein-Ligand Complexes from Standard Protein Data Bank File Input. Protein Eng. 1995, 8, 127–134. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Warren, L.; DeLano, P.D. PyMOL: An Open-Source Molecular Graphics Tool. Ccp4.Ac.Uk. 2002. Available online: http://legacy.ccp4.ac.uk/newsletters/newsletter40/11_pymol.pdf (accessed on 26 August 2022).
- Berendsen, H.J.C.; van der Spoel, D.; van Drunen, R. GROMACS: A Message-Passing Parallel Molecular Dynamics Implementation. Comput. Phys. Commun. 1995, 91, 43–56. [Google Scholar] [CrossRef]
- Hornak, V.; Abel, R.; Okur, A.; Strockbine, B.; Roitberg, A.; Simmerling, C. Comparison of Multiple Amber Force Fields and Development of Improved Protein Backbone Parameters. Proteins Struct. Funct. Bioinforma. 2006, 65, 712–725. [Google Scholar] [CrossRef] [Green Version]
- Sousa Da Silva, A.W.; Vranken, W.F. ACPYPE—AnteChamber PYthon Parser InterfacE. BMC Res. Notes 2012. [Google Scholar] [CrossRef] [PubMed]
- Kumari, R.; Kumar, R.; Lynn, A. G_mmpbsa—A GROMACS Tool for High-Throughput MM-PBSA Calculations. J. Chem. Inf. Model. 2014, 54, 1951–1962. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Cheng, F.; Chen, L.; Du, Z.; Li, W.; Liu, G.; Lee, P.W.; Tang, Y. In Silico Prediction of Chemical Ames Mutagenicity. J. Chem. Inf. Model. 2012, 52, 2840–2847. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Shen, J.; Yu, Y.; Li, W.; Liu, G.; Lee, P.W.; Tang, Y. In Silico Prediction of Tetrahymena Pyriformis Toxicity for Diverse Industrial Chemicals with Substructure Pattern Recognition and Machine Learning Methods. Chemosphere 2011, 82, 1636–1643. [Google Scholar] [CrossRef]
- Liao, S.-Y.; Mo, G.-Q.; Chen, J.-C.; Zheng, K.-C. Exploration of the Binding Mode between (−)-Zampanolide and Tubulin Using Docking and Molecular Dynamics Simulation. J. Mol. Model. 2014, 20, 2070. [Google Scholar] [CrossRef]
- Qais, F.A.; Sarwar, T.; Ahmad, I.; Khan, R.A.; Shahzad, S.A.; Husain, F.M. Glyburide Inhibits Non-Enzymatic Glycation of HSA: An Approach for the Management of AGEs Associated Diabetic Complications. Int. J. Biol. Macromol. 2021, 169, 143–152. [Google Scholar] [CrossRef]
- Fouedjou, R.T.; Chtita, S.; Bakhouch, M.; Belaidi, S.; Ouassaf, M.; Djoumbissie, L.A.; Tapondjou, L.A.; Abul Qais, F. Cameroonian Medicinal Plants as Potential Candidates of SARS-CoV-2 Inhibitors. J. Biomol. Struct. Dyn. 2021, 1–15. [Google Scholar] [CrossRef]
- Siddiqui, S.; Ameen, F.; Jahan, I.; Nayeem, S.M.; Tabish, M. A Comprehensive Spectroscopic and Computational Investigation on the Binding of the Anti-Asthmatic Drug Triamcinolone with Serum Albumin. New J. Chem. 2019, 43, 4137–4151. [Google Scholar] [CrossRef]
- Ding, K.; Zhang, H.; Wang, H.; Lv, X.; Pan, L.; Zhang, W.; Zhuang, S. Atomic-Scale Investigation of the Interactions between Tetrabromobisphenol A, Tetrabromobisphenol S and Bovine Trypsin by Spectroscopies and Molecular Dynamics Simulations. J. Hazard. Mater. 2015, 299, 486–494. [Google Scholar] [CrossRef]
- Rath, B.; Abul Qais, F.; Patro, R.; Mohapatra, S.; Sharma, T. Design, Synthesis and Molecular Modeling Studies of Novel Mesalamine Linked Coumarin for Treatment of Inflammatory Bowel Disease. Bioorg. Med. Chem. Lett. 2021, 128029. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, S.; Ameen, F.; Kausar, T.; Nayeem, S.M.; Ur Rehman, S.; Tabish, M. Biophysical Insight into the Binding Mechanism of Doxofylline to Bovine Serum Albumin: An in Vitro and in Silico Approach. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2021, 249, 119296. [Google Scholar] [CrossRef] [PubMed]




| Compound Names | Micro Constituent | Formula | Reference |
|---|---|---|---|
| Rutin | Flavonoid | C27H30O16 | [13] |
| Luteolin_7_O_glucoside | Flavonoid | C21H20O11 | [14] |
| Verbascoside | Phenolic compound | C29H36O15 | [13] |
| Maslinic acid | Triterpenoid | C30H48O4 | [15] |
| Oleuropein | Secoiridoid | C25H32O13 | [1] |
| Lucidumoside C | Secoiridoid | C27H36O14 | [14] |
| Ursolic acid | Triterpenoid | C30H48O3 | [13] |
| Oleanolic acid | Triterpenoid | C30H48O3 | [13] |
| Uvaol | Phenolic compound | C30H50O2 | [15] |
| Betulinic acid | Triterpenoid | C30H48O3 | [16] |
| Oleoside | Secoiridoid | C16H22O11 | [13] |
| Elenolic acid glucoside | Secoiridoid | C17H25O11 | [13] |
| Loganic acid | Phenolic compound | C16H24O10 | [17] |
| Diosmetin | Flavone glycoside | C21H13O11 | [18] |
| Apigenin | Flavonoid | C15H10O5 | [13,14] |
| Secologanin | Phenolic compound | C17H24O10 | [17] |
| Ferulic acid | Phenolic compound | C10H10O4 | [19] |
| Hydroxytyrosol | Phenolic compound | C8H10O3 | [13] |
| Gallic acid | Phenolic compound | C7H6O5 | [20] |
| Coumaric acid | Phenolic compound | C9H8O3 | [21] |
| Homovanillyl alcohol | Phenolic compound | C9H12O3 | [13] |
| Cinnamic acid | Phenolic compound | C9H8O2 | [13] |
| Tyrosol | Phenolic compound | C8H10O2 | [22] |
| Compound Names | AMES Toxicity | hERG-I Inhibitor | Hepato-Toxicity | Skin Sensitisation | T. pyriformis Toxicity (log µg/L) | Oral Rat Acute Toxicity (mol/kg) |
|---|---|---|---|---|---|---|
| Rutin | No | No | No | No | 0.285 | 2.491 |
| Verbascoside | No | No | No | No | 0.285 | 2.527 |
| Luteolin_7_O_glucoside | No | No | No | No | 0.285 | 2.547 |
| Ursolic acid | No | No | Yes | No | 0.285 | 2.346 |
| Oleanolic Acid | No | No | Yes | No | 0.285 | 2.349 |
| Oleuropein | No | No | No | No | 0.285 | 2.862 |
| Maslinic acid | No | No | Yes | No | 0.285 | 2.516 |
| Uvaol | No | No | Yes | No | 0.376 | 2.646 |
| Betulinic acid | No | No | Yes | No | 0.285 | 2.256 |
| Apigenin | No | No | No | No | 0.38 | 2.45 |
| Loganic acid | No | No | No | No | 0.285 | 1.995 |
| Oleoside | No | No | No | No | 0.285 | 2.389 |
| Diosmetin | No | No | No | No | 0.336 | 2.338 |
| Elenolic acid glucoside | No | No | No | No | 0.285 | 2.35 |
| Lucidumoside C | No | No | No | No | 0.285 | 3.068 |
| Secologanin | No | No | No | No | 0.285 | 2.031 |
| Coumaric acid | No | No | No | No | 0.319 | 2.155 |
| Cinnamic acid | No | No | No | No | 0.247 | 2.094 |
| Ferulic acid | No | No | No | No | 0.271 | 2.282 |
| Hydroxytyrosol | Yes | No | No | No | −0.128 | 1.858 |
| Gallic acid | No | No | No | No | 0.285 | 2.218 |
| Tyrosol | No | No | No | Yes | −0.244 | 1.861 |
| Homovanillyl alcohol | No | No | No | No | −0.059 | 1.891 |
| 1/total | 0/total | 5/total | 1/total |
| Compound Names | Absorption | Distribution | Metabolism | Excretion | |
|---|---|---|---|---|---|
| Water Solubility (log mol/L) | Intestinal Absorption (%) | Fraction Unbound (Fu) | CYP3A4 Inhibitor | Total Clearance (log mL/min/kg) | |
| Rutin | −2.892 | 3.446 | 0.187 | No | −0.369 |
| Verbascoside | −2.906 | 32.119 | 0.269 | No | 0.479 |
| Luteolin_7_O_glucoside | −2.716 | 37.556 | 0.224 | No | 0.478 |
| Ursolic acid | −3.072 | 100 | 0 | No | 0.083 |
| Oleanolic Acid | −3.074 | 99.931 | 0 | No | 0.285 |
| Oleuropein | −2.722 | 44.206 | 0.485 | No | 1.176 |
| Maslinic acid | −3.042 | 100 | 0.033 | No | −0.071 |
| Uvaol | −5.947 | 92.819 | 0 | No | 0.206 |
| Betulinic acid | −3.122 | 99.763 | 0.018 | No | 0.116 |
| Apigenin | −3.329 | 93.25 | 0.147 | No | 0.566 |
| Loganic acid | −2.365 | 16.515 | 0.641 | No | 1.225 |
| Oleoside | −2.604 | 0 | 0.567 | No | 1.395 |
| Diosmetin | −3.238 | 79.898 | 0.068 | No | 0.598 |
| Elenolic acid glucoside | −2.517 | 20.4 | 0.606 | No | 1.482 |
| Lucidumoside C | −2.833 | 41.241 | 0.418 | No | 1.146 |
| Secologanin | −2.676 | 40.54 | 0.611 | No | 1.585 |
| Coumaric acid | −2.378 | 93.494 | 0.428 | No | 0.662 |
| Cinnamic acid | −2.608 | 94.833 | 0.38 | No | 0.781 |
| Ferulic acid | −2.817 | 93.685 | 0.343 | No | 0.623 |
| Hydroxytyrosol | −1.139 | 72.809 | 0.593 | No | 0.23 |
| Gallic acid | −2.56 | 43.374 | 0.617 | No | 0.518 |
| Tyrosol | −1.146 | 85.255 | 0.485 | No | 0.283 |
| Homovanillyl alcohol | −1.433 | 84.608 | 0.395 | No | 0.305 |
| Compound Name | Pyruvate Kinase Inhibition | Antineoplastic | ||
|---|---|---|---|---|
| Pa | Pi | Pa | Pi | |
| Rutin | 0.229 | 0.009 | 0.849 | 0.007 |
| Luteolin_7_O_glucoside | 0.680 | 0.002 | 0.830 | 0.009 |
| Verbascoside | 0.115 | 0.017 | 0.814 | 0.010 |
| Maslinic | -- | -- | 0.867 | 0.005 |
| Oleuropein | -- | -- | 0.387 | 0.109 |
| Lucidumoside C | -- | -- | 0.674 | 0.030 |
| Ursolic acid | -- | -- | 0.857 | 0.006 |
| Oleanolic acid | -- | -- | 0.876 | 0.005 |
| Uvaol | -- | -- | 0.907 | 0.005 |
| Betulinic acid | -- | -- | 0.925 | 0.005 |
| Oleoside | 0.091 | 0.025 | 0.796 | 0.012 |
| Elenolic acid glucoside | 0.063 | 0.050 | 0.795 | 0.012 |
| Loganic acid | 0.071 | 0.040 | 0.817 | 0.010 |
| Diosmetin | 0.135 | 0.014 | 0.791 | 0.013 |
| Apigenin | 0.280 | 0.007 | 0.774 | 0.015 |
| Secologanin | -- | -- | 0.500 | 0.071 |
| Ferulic acid | 0.080 | 0.031 | 0.601 | 0.045 |
| Hydroxytyrosol | 0.119 | 0.016 | -- | -- |
| Gallic acid | 0.124 | 0.016 | 0.313 | 0.145 |
| Coumaric acid | 0.163 | 0.011 | 0.520 | 0.065 |
| Homovanillyl alcohol | 0.070 | 0.040 | 0.321 | 0.140 |
| Cinnamic acid | 0.141 | 0.013 | 0.455 | 0.085 |
| Tyrosol | 0.132 | 0.014 | -- | -- |
| Compound Names | PubChem CID | Binding Energy (kcal/mol) | Ki (µmol/L) | Binding Site |
|---|---|---|---|---|
| Rutin | 5280805 | −10 | 0.046 | CS |
| Verbascoside | 5281800 | −9.8 | 0.064 | CS |
| Luteolin_7_O_glucoside | 5280637 | −8.9 | 0.296 | CS |
| Ursolic acid | 64945 | −8.9 | 0.296 | OS |
| Oleanolic Acid | 10494 | −8.8 | 0.351 | CS |
| Oleuropein | 5281544 | −8.8 | 0.351 | CS |
| Maslinic | 73659 | −8.7 | 0.415 | OS |
| Uvaol | 92802 | −8.7 | 0.415 | OS |
| Betulinic acid | 64971 | −8.4 | 0.690 | OS |
| Apigenin | 5280443 | −8.1 | 1.145 | OS |
| Loganic acid | 89640 | −8 | 1.356 | CS |
| Oleoside | 101042548 | −8 | 1.356 | CS |
| Diosmetin | 5281612 | −9 | 0.250 | CS |
| Elenolic acid glucoside | 10692563 | −7.6 | 2.665 | CS |
| Lucidumoside C | 10793430 | −7.5 | 3.155 | CS |
| Secologanin | 161276 | −7.1 | 6.201 | CS |
| Coumaric acid | 637542 | −6.9 | 8.693 | AS |
| Cinnamic acid | 444539 | −6.4 | 20.226 | CS |
| Ferulic acid | 445858 | −6.3 | 23.947 | OS |
| Hydroxytyrosol | 82755 | −6.3 | 23.947 | AS |
| Gallic acid | 370 | −6 | 39.746 | CS |
| Tyrosol | 10393 | −6 | 39.746 | AS |
| Homovanillyl alcohol | 16928 | −5.7 | 65.968 | CS |
| Ligands | |||
|---|---|---|---|
| Verbascoside | Rutin | Luteolin_7_O_Glucoside | |
| ΔEvdW | −47.63 ± 6.78 | −35.32 ± 2.91 | −40.88 ± 5.98 |
| ΔEele | −39.57 ± 6.32 | −40.47 ± 6.57 | −26.22 ± 10.80 |
| ΔEPSE | 85.42 ± 11.35 | 77.30 ± 12.81 | 56.68 ± 17.23 |
| ΔESSASA | −5.98 ± 0.39 | −4.72 ± 0.44 | −4.57 ± 0.50 |
| ΔEBE | −7.76 ± 5.95 | −3.22 ± 3.22 | −15.00 ± 5.58 |
| Luteolin_7_O_Glucoside | Rutin | Verbascoside | |||
|---|---|---|---|---|---|
| Residues | Etotal | Residues | Etotal | Residues | Etotal |
| Thr50 | −0.71 ± 0.04 | Thr328 | −0.68 ± 0.06 | Thr50 | −0.72 ± 0.02 |
| Pro53 | −1.96 ± 0.04 | Gln329 | −0.80 ± 0.11 | Pro53 | −0.68 ± 0.02 |
| Leu74 | −0.63 ± 0.04 | Ile335 | −1.38 ± 0.07 | Ser77 | −0.59 ± 0.08 |
| His78 | −1.87 ± 0.09 | Gly363 | −0.61 ± 0.05 | His78 | −0.54 ± 0.05 |
| Gly79 | −0.52 ± 0.04 | Glu364 | −0.55 ± 0.25 | Asp113 | −0.81 ± 0.17 |
| Tyr83 | −2.20 ± 0.12 | Thr114 | −0.73 ± 0.06 | ||
| Ala366 | −0.77 ± 0.05 | Glu118 | −1.11 ± 0.05 | ||
| Val176 | −0.60 ± 0.03 | ||||
| Asp177 | −0.96 ± 0.05 | ||||
| Val209 | −0.88 ± 0.02 | ||||
| Glu332 | −0.56 ± 0.08 | ||||
| Ala366 | −0.59 ± 0.03 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qais, F.A.; Alomar, S.Y.; Imran, M.A.; Hashmi, M.A. In-Silico Analysis of Phytocompounds of Olea europaea as Potential Anti-Cancer Agents to Target PKM2 Protein. Molecules 2022, 27, 5793. https://doi.org/10.3390/molecules27185793
Qais FA, Alomar SY, Imran MA, Hashmi MA. In-Silico Analysis of Phytocompounds of Olea europaea as Potential Anti-Cancer Agents to Target PKM2 Protein. Molecules. 2022; 27(18):5793. https://doi.org/10.3390/molecules27185793
Chicago/Turabian StyleQais, Faizan Abul, Suliman Yousef Alomar, Mohammad Azhar Imran, and Md Amiruddin Hashmi. 2022. "In-Silico Analysis of Phytocompounds of Olea europaea as Potential Anti-Cancer Agents to Target PKM2 Protein" Molecules 27, no. 18: 5793. https://doi.org/10.3390/molecules27185793






