Ethidium Binding to Salmonella enterica ser. Typhimurium Cells and Salmon Sperm DNA
Abstract
:1. Introduction
2. Results
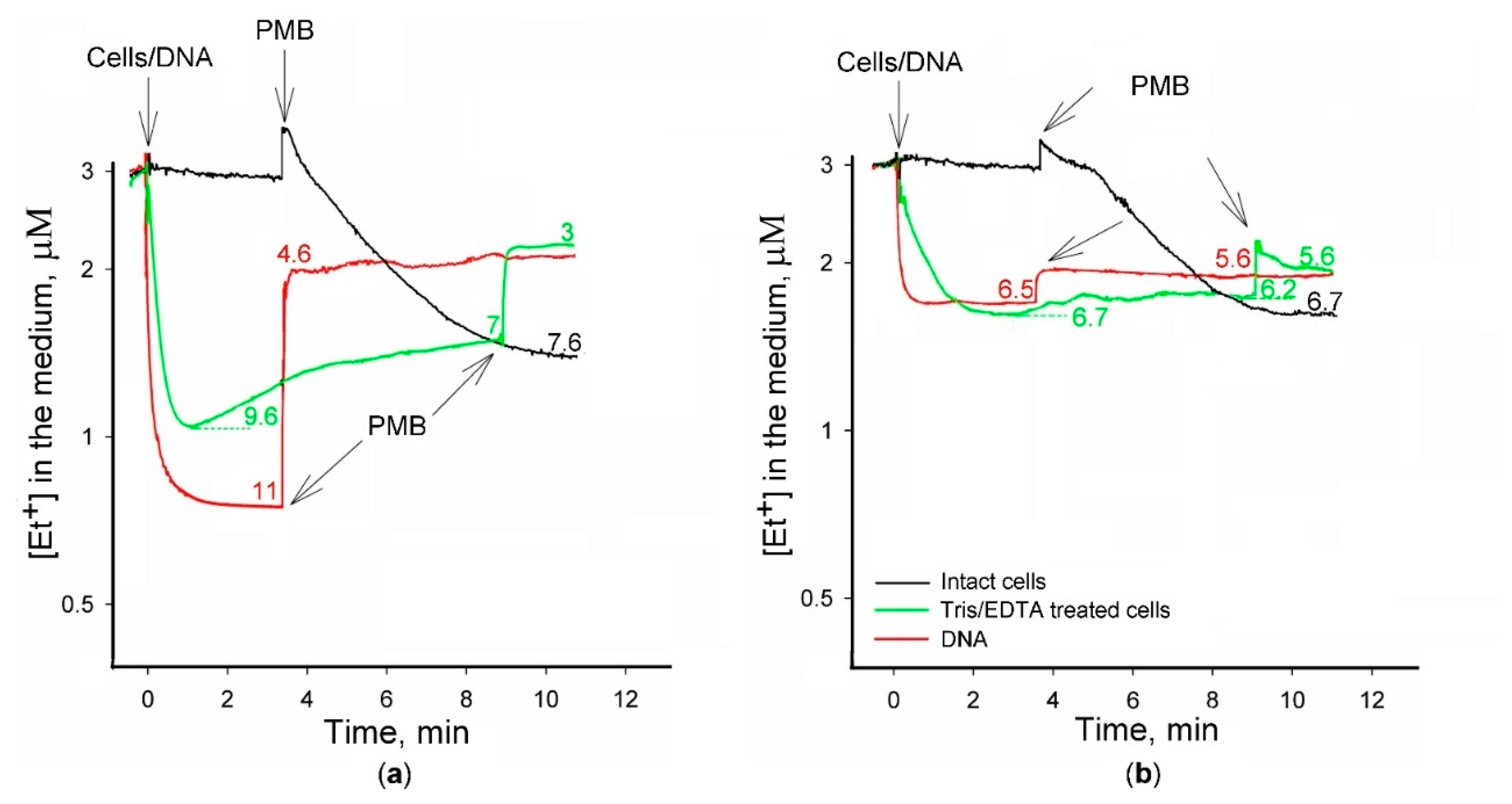
2.1. Simultaneous Measurements of Et+ and TPP+ Interaction with S. typhimurium Cells
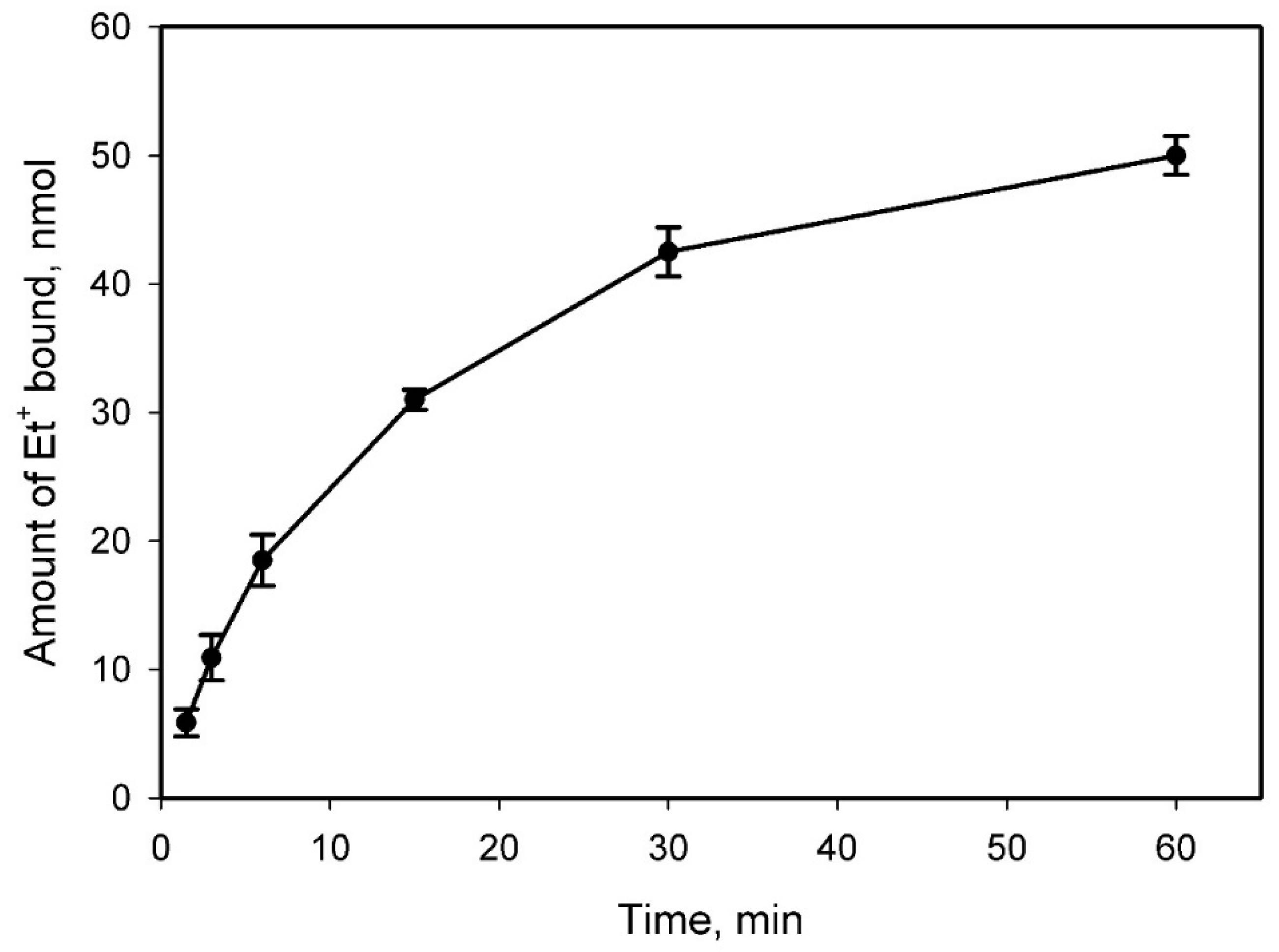
2.2. Et+ Binding to S. typhimurium Cells
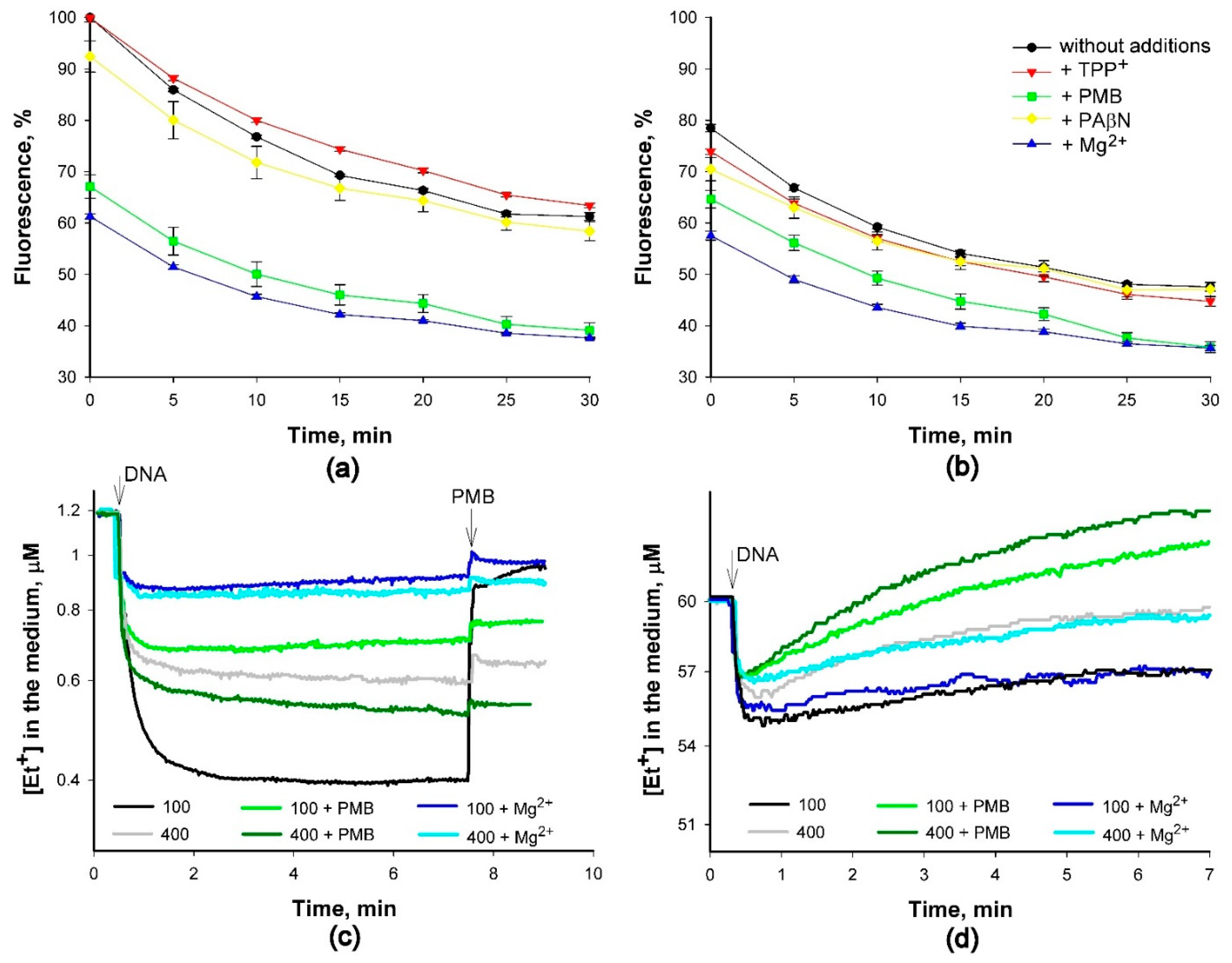
2.3. Binding of Et+ to DNA in Solutions of Various Composition
3. Discussion
4. Materials and Methods
4.1. Bacteria Cultivation and Preparation for Experiments
4.2. Potentiometric Measurements
4.3. Fluorescence Measurements
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Du, D.; Wang-Kan, X.; Neuberger, A.; van Veen, H.W.; Pos, K.M.; Piddock, L.J.V.; Luisi, B.F. Multidrug efflux pumps: Structure, function and regulation. Nat. Rev. Microbiol. 2018, 16, 523–539. [Google Scholar] [CrossRef]
- Wang, Z.; Fan, G.; Hryc, C.F.; Corey, F.; Blaza, J.N.; Serysheva, I.I.; Schmid, M.F.; Chiu, W.; Luisi, B.F.; Du, D. An allosteric transport mechanism for the AcrAB-TolC multidrug efflux pump. eLife 2017, 6, 1–19. [Google Scholar] [CrossRef]
- Gros, P.; Talbot, F.; Tang-Wai, D.; Bibi, E.; Kaback, H.R. Lipophilic cations: A group of model substrates for the multidrug-resistance transporter. Biochemistry 1992, 31, 1992–1998. [Google Scholar] [CrossRef]
- Blair, J.M.A.; Piddock, L.J.V. How to measure export via bacterial multidrug resistance efflux pumps. mBio 2016, 7, e00840-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daugelavicius, R.; Buivydas, A.; Sencilo, A.; Bamford, D.H. Assessment of the activity of RND-type multidrug efflux pumps in Pseudomonas aeruginosa using tetraphenylphosphonium ions. Int. J. Antimicrob. Agents 2010, 36, 234–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martins, A.; Spengler, G.; Rodrigues, L.; Rodrigues, G.; Viveiros, L.; Ramos, J.; Martins, M.; Couto, I.; Fanning, S.; Pages, J.-M.; et al. pH Modulation of efflux pump activity of multi-drug resistant Escherichia coli: Protection during its passage and eventual colonization of the colon. PLoS ONE 2009, 4, e6656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hockings, P.D.; Rogers, P.J. The measurement of transmembrane electrical potential with lipophilic cations. Biochim. Biophys. Acta 1996, 1282, 101–106. [Google Scholar] [CrossRef] [Green Version]
- Nicholls, D.G.; Ferguson, S. Bioenergetics, 4th ed.; Elsevier Ltd.: Amsterdam, The Netherlands, 2013. [Google Scholar]
- Severini, A.; Richard Morgan, A. An assay for proteinases and their inhibitors based on DNA/ethidium bromide fluorescence. Anal. Biochem. 1991, 193, 83–89. [Google Scholar] [CrossRef]
- Martins, A.; MacHado, L.; Costa, S.; Cerca, P.; Spengler, G.; Viveiros, M.; Amaral, L. Role of calcium in the efflux system of Escherichia coli. Int. J. Antimicrob. Agents 2011, 37, 410–414. [Google Scholar] [CrossRef]
- Amaral, L.; Cerca, P.; Spengler, G.; MacHado, L.; Martins, A.; Couto, I.; Viveiros, M.; Fanning, S.; Pages, J.M. Ethidium bromide efflux by Salmonella: Modulation by metabolic energy, pH, ions and phenothiazines. Int. J. Antimicrob. Agents 2011, 38, 140–145. [Google Scholar] [CrossRef] [Green Version]
- Anbazhagan, P.V.; Thavitiki, P.R.; Varra MAnnamalai, L.; Putturu, R.; Lakkineni, V.R.; Pesingi, P.K. Evaluation of efflux pump activity of multidrug-resistant Salmonella typhimurium isolated from poultry wet markets in India. Infect. Drug Resist. 2019, 12, 1081–1088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Babayan, A.; Nikaido, H. In Pseudomonas aeruginosa ethidium bromide does not induce its own degradation or the assembly of pumps involved in its efflux. Biochem. Biophys. Res. Commun. 2004, 324, 1065–1068. [Google Scholar] [CrossRef] [PubMed]
- Mikalayeva, V.; Sakalauskaitė, S.; Daugelavičius, R. Interaction of ethidium and tetraphenylphosphonium cations with Salmonella enterica cells. Medicina (Lithuania) 2017, 53, 122–130. [Google Scholar] [CrossRef]
- Rodrigues, L.; Wagner, D.; Viveiros, M.; Sampaio, D.; Couto, I.; Vavra, M.; Kern, W.V.; Amaral, L. Thioridazine and chlorpromazine inhibition of ethidium bromide efflux in Mycobacterium avium and Mycobacterium smegmatis. J. Antimicrob. Chemoth. 2008, 61, 1076–1082. [Google Scholar] [CrossRef]
- Daugelavicius, R.; Bakiene, E.; Bamford, D.H. Stages of polymyxin B interaction with the Escherichia coli cell envelope. Antimicrob. Agents Chem. 2000, 44, 2969–2978. [Google Scholar] [CrossRef] [Green Version]
- Sutkuvienė, S.; Mikalayeva, V.; Pavan, S.; Berti, F.; Daugelavičius, R. Evaluation of the Efficiency of Synthesized Efflux Pump Inhibitors on Salmonella enterica ser. Typhimurium Cells. Chem. Biol. Drug Des. 2013, 82, 438–445. [Google Scholar] [CrossRef]
- Xu, X.H.N.; Wan, Q.; Kyriacou, S.V.; Brownlow, W.J.; Nowak, M.E. Direct observation of substrate induction of resistance mechanism in Pseudomonas aeruginosa using single live cell imaging. Biochem. Biophys. Res. Commun. 2003, 305, 941–949. [Google Scholar] [CrossRef]
- Lepecq, J.B.; Paoletti, C. A fluorescent complex between ethidium bromide and nucleic acids: Physical-chemical characterization. J. Mol. Biol. 1967, 27, 87–106. [Google Scholar] [CrossRef]
- Hayashi, M.; Harada, Y. Direct observation of the reversible unwinding of a single DNA molecule caused by the intercalation of ethidium bromide. Nucleic Acids Res. 2007, 35, e125. [Google Scholar] [CrossRef] [PubMed]
- Heller, D.P.; Greenstock, C.L. Fluorescence lifetime analysis of DNA intercalated ethidium bromide and quenching by free dye. Biophys. Chem. 1994, 50, 305–312. [Google Scholar] [CrossRef]
- Neidhardt, F.C.; Curtiss III, R.; Ingraham, J.L.; Lin, E.C.C.; Low, K.B.; Magasanik, B.; Reznikoff, W.S.; Riley, M.; Schaechter, M.; Umbarger, H.E. Escherichia coli and Salmonella: Cellular and Molecular Biology, 2nd ed.; ASM Press: Washington, DC, USA, 1996. [Google Scholar]
- McClelland, M.; Sanderson, K.E.; Spieth, J.; Thomson, N.R.; Pickard, D.; Wain, J.; Churcher, C.; Mungall, K.L.; Bentley, S.D.; Holden, M.T.G.; et al. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 2001, 413, 852–856. [Google Scholar] [CrossRef] [Green Version]
- Goeden, D.J.R.; Shao, B.M.Y.; Glasner, D.; Rode, C.K.; Mayhew, G.F.; Gregor, J.; Davis, N.W.; Kirkpatrick, H.A.; Frederick, M.A.; Blattner, R.; et al. The Complete Genome Sequence of Escherichia coli K-12. Science 1997, 277, 1453–1462. [Google Scholar] [CrossRef] [Green Version]
- Martz, W.W. The Interaction of Ethidium Bromide with Nucleic Acids, Ecommons. 1971. Available online: https://ecommons.luc.edu/luc_diss/1149 (accessed on 12 April 2021).
- Vardevanyan, P.O.; Antonyan, A.P.; Parsadanyan, M.A.; Davtyan, H.G.; Karapetyan, A.T. The binding of ethidium bromide with DNA: Interaction with single- and double-stranded structures. Exp. Mol. Med. 2003, 35, 527–533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jernaes, M.W.; Steen, H.B. Staining of Escherichia coli for flow cytometry: Influx and efflux of ethidium bromide. Cytometry 1994, 17, 302–309. [Google Scholar] [CrossRef]
- Badrinarayanan, A.; Le, T.B.K.; Laub, M.T. Bacterial chromosome organization and segregation. Annu. Rev. Cell Dev. Biol. 2015, 31, 171–199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodrigues, L.; Ramos, J.; Couto, I.; Amaral, L.; Viveiros, M. Ethidium bromide transport across Mycobacterium smegmatis cell-wall : Correlation with antibiotic resistance. BMC Microbiol. 2011, 11, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marvin, H.J.; ter Beest, M.B.; Witholt, B. Release of outer membrane fragments from wild-type Escherichia coli and from several E. coli lipopolysaccharide mutants by EDTA and heat shock treatments. J. Bacteriol. 1989, 171, 5262–5267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daugelavicius, R.; Bamford, J.K.; Bamford, D.H. Changes in host cell energetics in response to bacteriophage PRD1 DNA entry. J. Bacteriol. 1997, 179, 5203–5210. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sakalauskaitė, S.; Mikalayeva, V.; Daugelavičius, R. Ethidium Binding to Salmonella enterica ser. Typhimurium Cells and Salmon Sperm DNA. Molecules 2021, 26, 3386. https://doi.org/10.3390/molecules26113386
Sakalauskaitė S, Mikalayeva V, Daugelavičius R. Ethidium Binding to Salmonella enterica ser. Typhimurium Cells and Salmon Sperm DNA. Molecules. 2021; 26(11):3386. https://doi.org/10.3390/molecules26113386
Chicago/Turabian StyleSakalauskaitė, Sandra, Valeryia Mikalayeva, and Rimantas Daugelavičius. 2021. "Ethidium Binding to Salmonella enterica ser. Typhimurium Cells and Salmon Sperm DNA" Molecules 26, no. 11: 3386. https://doi.org/10.3390/molecules26113386





