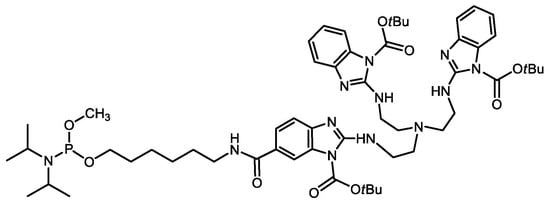
A Trisbenzimidazole Phosphoramidite Building Block Enables High-Yielding Syntheses of RNA-Cleaving Oligonucleotide Conjugates
Abstract
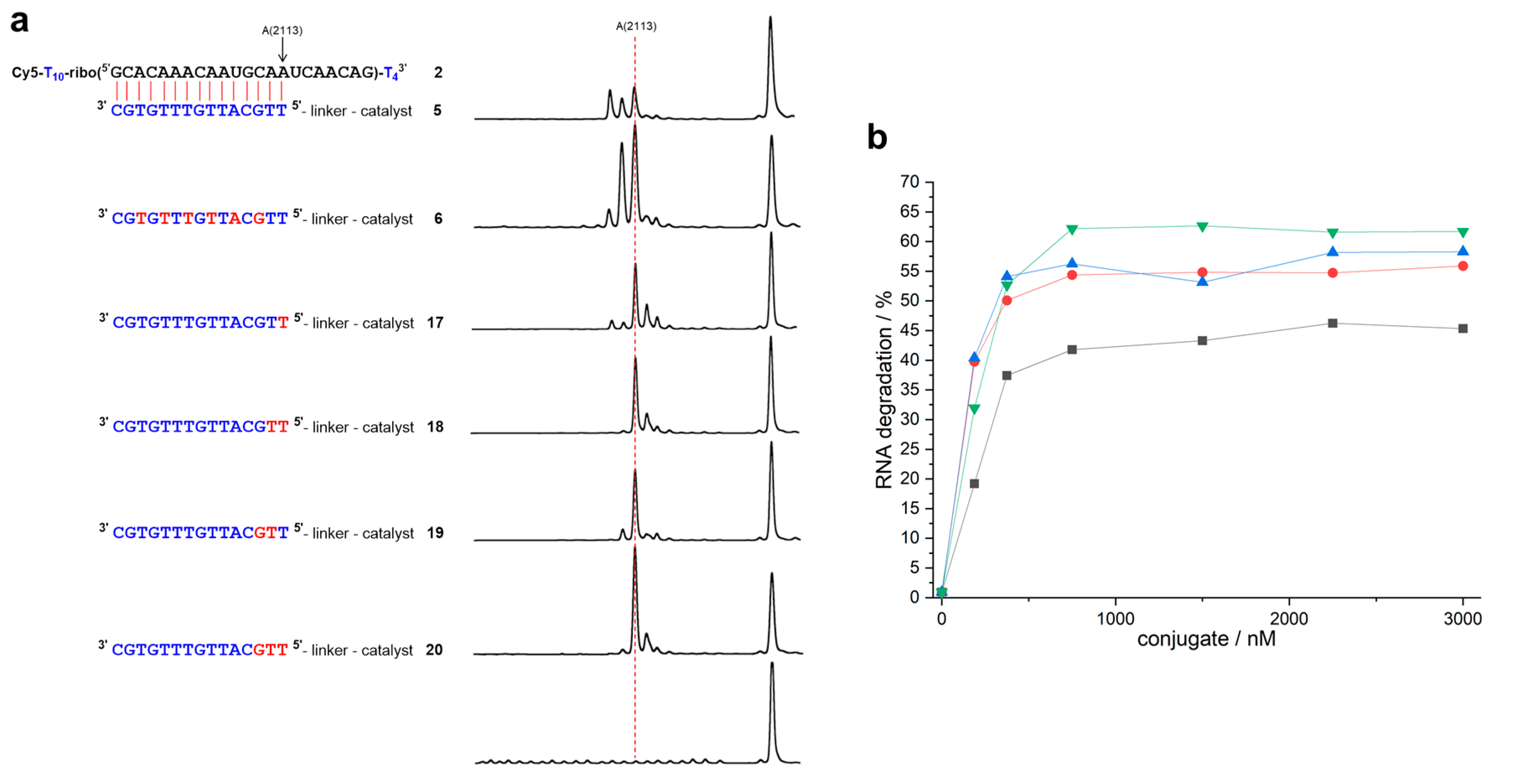
:1. Introduction
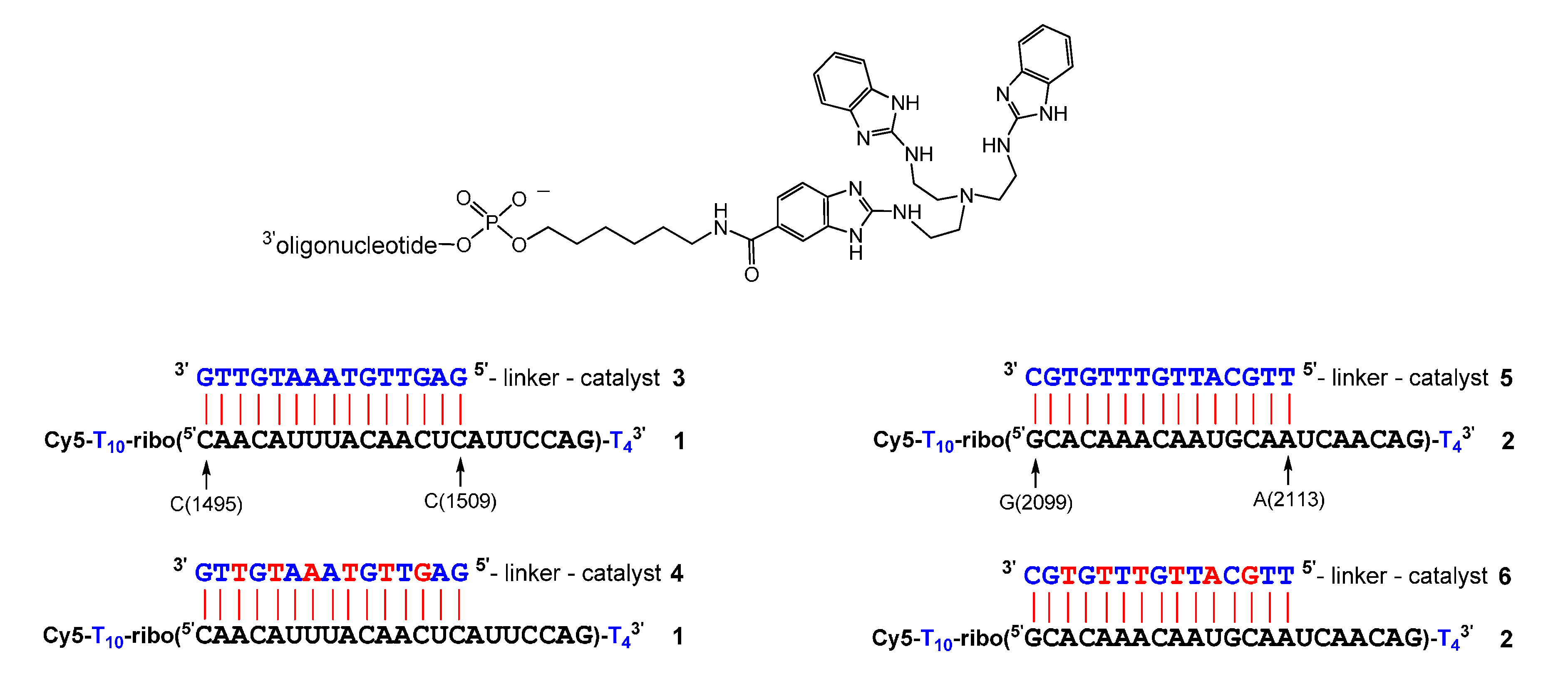
2. Results
3. Discussion
4. Materials and Methods
4.1. General
4.2. Synthesis, Purification, Quantification and Analysis of Conjugates
4.2.1. Manual Phosphoramidite Coupling
4.2.2. Isolation and Purification
4.2.3. Quantification
4.2.4. Mass spectrometry
4.2.5. Cleavage Studies
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Furdon, P.J.; Dominski, Z.; Ryszard Kole, R. RNase H cleavage of RNA hybridized to oligonudeotides containing methylphosphonate, phosphorothioate and phosphodiester bonds. Nucleic Acids Res. 1989, 17, 9193–9204. [Google Scholar] [CrossRef] [Green Version]
- Agrawal, S.; Mayrand, S.H.; Zamecnik, P.C.; Pederson, T. Site-specific excision from RNA by RNase H and mixed-phosphate-backbone oligodeoxynucleotides. Proc. Natl. Acad. Sci. U.S.A. 1990, 87, 1401–1405. [Google Scholar] [CrossRef] [Green Version]
- Hall, J.; Hüsken, D.; Pieles, U.; Moser, H.E.; Häner, R. Efficient sequence-specific cleavage of RNA using novel europium complexes conjugated to oligonucleotides. Chem. Biol. 1994, 1, 185–190. [Google Scholar] [CrossRef]
- Bashkin, J.K.; Frolova, E.I.; Sampath, U. Sequence-specific cleavage of HIV mRNA by a ribozyme mimic. J. Am. Chem. Soc. 1994, 116, 5981–5982. [Google Scholar] [CrossRef]
- Matsumura, K.; Endo, M.; Komiyama, M. Lanthanide complex–oligo-DNA hybrid for sequence-selective hydrolysis of RNA. J. Chem. Soc. Chem. Commun. 1994, 2019–2020. [Google Scholar] [CrossRef]
- Magda, D.; Miller, R.A.; Sessler, J.L.; Iverson, B.L. Site-specific hydrolysis of RNA by europium(III) texaphyrin conjugated to a synthetic oligodeoxyribonucleotide. J. Am. Chem. Soc. 1994, 116, 7439–7440. [Google Scholar] [CrossRef]
- Hovinen, J.; Guzaev, A.; Azhayeva, E.; Azhayev, A.; Lönnberg, H. Imidazole Tethered Oligodeoxyribonucleotides: Synthesis and RNA Cleaving Activity. J. Org. Chem. 1995, 60, 2205–2209. [Google Scholar] [CrossRef]
- Trawick, B.N.; Daniher, A.T.; Bashkin, J.K. Inorganic mimics of ribonucleases and ribozymes: From random cleavage to sequence-specific chemistry to catalytic antisense drugs. Chem. Rev. 1998, 98, 939–960. [Google Scholar] [CrossRef]
- Häner, R. Artificial ribonucleases. Chimia 2001, 55, 1035–1037. [Google Scholar]
- Komiyama, M.; Sumaoka, J.; Kuzuya, A.; Yamamoto, Y. Sequence-selective artificial ribonucleases. Methods Enzymol. 2001, 341, 455–468. [Google Scholar]
- Morrow, J.R.; Iranzo, O. Synthetic metallonucleases for RNA cleavage. Curr. Opin. Chem. Biol. 2004, 8, 192–200. [Google Scholar] [CrossRef]
- Niittymäki, T.; Lönnberg, H. Artificial ribonucleases. Org. Biomol. Chem. 2006, 4, 15–25. [Google Scholar] [CrossRef]
- Lönnberg, H. Cleavage of RNA phosphodiester bonds by small molecular entities: A mechanistic insight. Org. Biomol. Chem. 2011, 9, 1687–1703. [Google Scholar] [CrossRef]
- Monia, B.P.; Lesnik, E.A.; Gonzalez, C.; Lima, W.F.; McGee, D.; Guinosso, C.J.; Kawasaki, A.M.; Cook, P.D.; Freier, S.M. Evaluation of 2‘-Modified Oligonucleotides Containing 2’-Deoxy Gaps as Antisense Inhibitors of Gene Expression. J. Biol. Chem. 1993, 268, 14514–14522. [Google Scholar]
- Jepsen, J.S.; Wengel, J. LNA-antisense rivals siRNA for gene silencing. Curr. Opin. Drug Discov. Devel. 2004, 7, 188–194. [Google Scholar]
- Elbashir, S.M.; Harborth, J.; Lendeckel, W.; Yalcin, A.; Weber, K.; Tuschl, T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001, 411, 494–498. [Google Scholar] [CrossRef]
- Murtola, M.; Wenska, M.; Strömberg, R. PNAzymes that are artificial RNA restriction enzymes. J. Am. Chem. Soc. 2010, 132, 8984–8990. [Google Scholar] [CrossRef]
- Ghidini, A.; Steunenberg, P.; Murtola, M.; Strömberg, R. Synthesis of PNA Oligoether Conjugates. Molecules 2014, 19, 3135–3148. [Google Scholar] [CrossRef] [Green Version]
- Ghidini, A.; Murtola, M.; Strömberg, R. Oligonucleotide based artificial ribonucleases (OBANs). In DNA in supramolecular chemistry and nanotechnology; Stulz, E., Clever, G.H., Eds.; John Wiley: Chichester, UK, 2015; pp. 158–171. ISBN 9781118696880. [Google Scholar]
- Ghidini, A.; Murtola, M.; Strömberg, R. Influence of conjugation and other structural changes on the activity of Cu2+ based PNAzymes. Org. Biomol. Chem. 2016, 14, 2768–2773. [Google Scholar] [CrossRef]
- Murtola, M.; Ghidini, A.; Virta, P.; Strömberg, R. Zinc Ion-Dependent Peptide Nucleic Acid-Based Artificial Enzyme that Cleaves RNA—Bulge Size and Sequence Dependence. Molecules 2017, 22, 1856. [Google Scholar] [CrossRef] [Green Version]
- Luige, O.; Murtola, M.; Ghidini, A.; Strömberg, R. Further Probing of Cu2+-Dependent PNAzymes Acting as Artificial RNA Restriction Enzymes. Molecules 2019, 24, 672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuzuya, A.; Machida, K.; Shi, Y.; Tanaka, K.; Komiyama, M. Site-Selective RNA Activation by Acridine-Modified Oligodeoxynucleotides in Metal-Ion Catalyzed Hydrolysis: A Comprehensive Study. ACS Omega 2017, 2, 5370–5377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salvio, R.; Volpi, S.; Folcarelli, T.; Casnati, A.; Cacciapaglia, R. A calix[4]arene with acylguanidine units as an efficient catalyst for phosphodiester bond cleavage in RNA and DNA model compounds. Org. Biomol. Chem. 2019, 17, 7482–7492. [Google Scholar] [CrossRef] [PubMed]
- Czescik, J.; Zamolo, S.; Darbre, T.; Mancin, F.; Scrimin, P. Factors Influencing the Activity of Nanozymes in the Cleavage of an RNA Model Substrate. Molecules 2019, 24, 2814. [Google Scholar] [CrossRef] [Green Version]
- Niittymäki, T.; Virta, P.; Ketomäki, K.; Lönnberg, H. Di(azacrown) Conjugates of 2‘-O-Methyl Oligoribonucleotides as Sequence-Selective Artificial Ribonucleases. Bioconjugate Chem. 2007, 18, 1583–1592. [Google Scholar] [CrossRef]
- Gaglione, M.; Milano, G.; Chambery, A.; Moggio, L.; Romanelli, A.; Messere, A. PNA-based artificial nucleases as antisense and anti-miRNA oligonucleotide agents. Mol. BioSyst. 2011, 7, 2490–2499. [Google Scholar] [CrossRef] [Green Version]
- Mironova, N.L.; Pyshnyi, D.V.; Shtadler, D.V.; Fedorova, A.A.; Vlassov, V.V.; Zenkova, M.A. RNase T1 mimicking artificial ribonuclease. Nucleic Acids Res. 2007, 35, 2356–2367. [Google Scholar] [CrossRef] [Green Version]
- Williams, A.; Staroseletz, Y.; Zenkova, M.A.; Jeannin, L.; Aojula, H.; Bichenkova, E.V. Peptidyl−Oligonucleotide Conjugates Demonstrate Efficient Cleavage of RNA in a Sequence-Specific Manner. Bioconjugate Chem. 2015, 26, 1129–1143. [Google Scholar] [CrossRef]
- Patutina, O.A.; Bichenkova, E.V.; Miroshnichenko, S.K.; Mironova, N.L.; Trivoluzzi, L.T.; Burusco, K.K.; Bryce, R.A.; Vlassov, V.V.; Zenkova, M.A. Novel peptide-oligonucleotide bioconjugates that silence miR-21 in lymphosarcoma cells. Biomaterials 2017, 122, 163–178. [Google Scholar] [CrossRef] [Green Version]
- Patutina, O.A.; Bazhenov, M.A.; Miroshnichenko, S.K.; Mironova, N.L.; Pyshnyi, D.V.; Vlassov, V.V.; Zenkova, M.A. Peptide-oligonucleotide conjugates exhibiting pyrimidine-X cleavage specificity efficiently silence miRNA target acting synergistically with RNase H. Sci. Rep. 2018, 8, 14990. [Google Scholar] [CrossRef] [Green Version]
- Pavlova, A.S.; Ogurtsova, P.A.; Koroleva, L.S.; Serpokrylova, I.Y.; Lomzov, A.A.; Pyshnaya, I.A.; Silnikov, V.N.; Pyshnyi, D.V. Novel Bisimidazole-Containing Peptidomimetic Molecules for Мetal-Independent RNA Cleavage: Synthesis and Solid-Phase Screening Method. Russ. J. Bioorg. Chem. 2019, 45, 813–824. [Google Scholar] [CrossRef]
- Trepanier, J.B.; Tanner, J.E.; Alfieri, C. Reduction in intracellular HCV RNA and virus protein expression in human hepatoma cells following treatment with 2-O-methyl-modified anti-core deoxyribozyme. Virology 2008, 377, 339–344. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Liu, E.; Lam, C.H.; Perrin, D.M. A densely modified M2+-independent DNAzyme that cleaves RNA efficiently with multiple catalytic turnover. Chem. Sci. 2018, 9, 1813–1821. [Google Scholar] [CrossRef] [PubMed]
- Scheffer, U.; Strick, A.; Ludwig, V.; Peter, S.; Kalden, E.; Göbel, M.W. Metal-free catalysts for the hydrolysis of RNA derived from guanidines, 2-aminopyridines, and 2-aminobenzimidazoles. J. Am. Chem. Soc. 2005, 127, 2211–2217. [Google Scholar] [CrossRef]
- Gnaccarini, C.; Peter, S.; Scheffer, U.; Vonhoff, S.; Klussmann, S.; Göbel, M.W. Site-specific cleavage of RNA by a metal-free artificial nuclease attached to antisense oligonucleotides. J. Am. Chem. Soc. 2006, 128, 8063–8067. [Google Scholar] [CrossRef] [PubMed]
- Danneberg, F.; Ghidini, A.; Dogandzhiyski, P.; Kalden, E.; Strömberg, R.; Göbel, M.W. Sequence-specific RNA cleavage by PNA conjugates of the metal-free artificial ribonuclease tris(2-aminobenzimidazole). Beilstein J. Org. Chem. 2015, 11, 493–498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dogandzhiyski, P.; Ghidini, A.; Danneberg, F.; Strömberg, R.; Göbel, M.W. Studies on tris(2-aminobenz- imidazole)-PNA based artificial nucleases: A comparison of two analytical techniques. Bioconjugate Chem. 2015, 26, 2514–2519. [Google Scholar] [CrossRef]
- Zellmann, F.; Thomas, L.; Scheffer, U.; Hartmann, R.K.; Göbel, M.W. Site-Specific Cleavage of RNAs Derived from the PIM1 3‘-UTR by a Metal-Free Artificial Ribonuclease. Molecules 2019, 24, 807. [Google Scholar] [CrossRef] [Green Version]
- Thomas, M.; Lange-Grünweller, K.; Weirauch, U.; Gutsch, D.; Aigner, A.; Grünweller, A.; Hartmann, R.K. The proto-oncogene Pim-1 is a target of miR-33a. Oncogene 2012, 31, 918–928. [Google Scholar] [CrossRef] [Green Version]
- Kierzek, E.; Ciesielska, A.; Pasternak, K.; Mathews, D.H.; Turner, D.H.; Kierzek, R. The influence of locked nucleic acid residues on the thermodynamic properties of 2’-O-methyl-RNA/RNA heteroduplexes. Nucleic Acids Res. 2005, 33, 5082–5093. [Google Scholar] [CrossRef] [Green Version]
- Nanodrop: Gray, D.M.; Hung, S.H.; Johnson, K.H. Absorption and Circular-Dichroism Spectroscopy of Nucleic-Acid Duplexes and Triplexes. Methods in Enzymology 1995, 246, 19. [Google Scholar]
Sample Availability: Samples of the compounds are not available from the authors. |

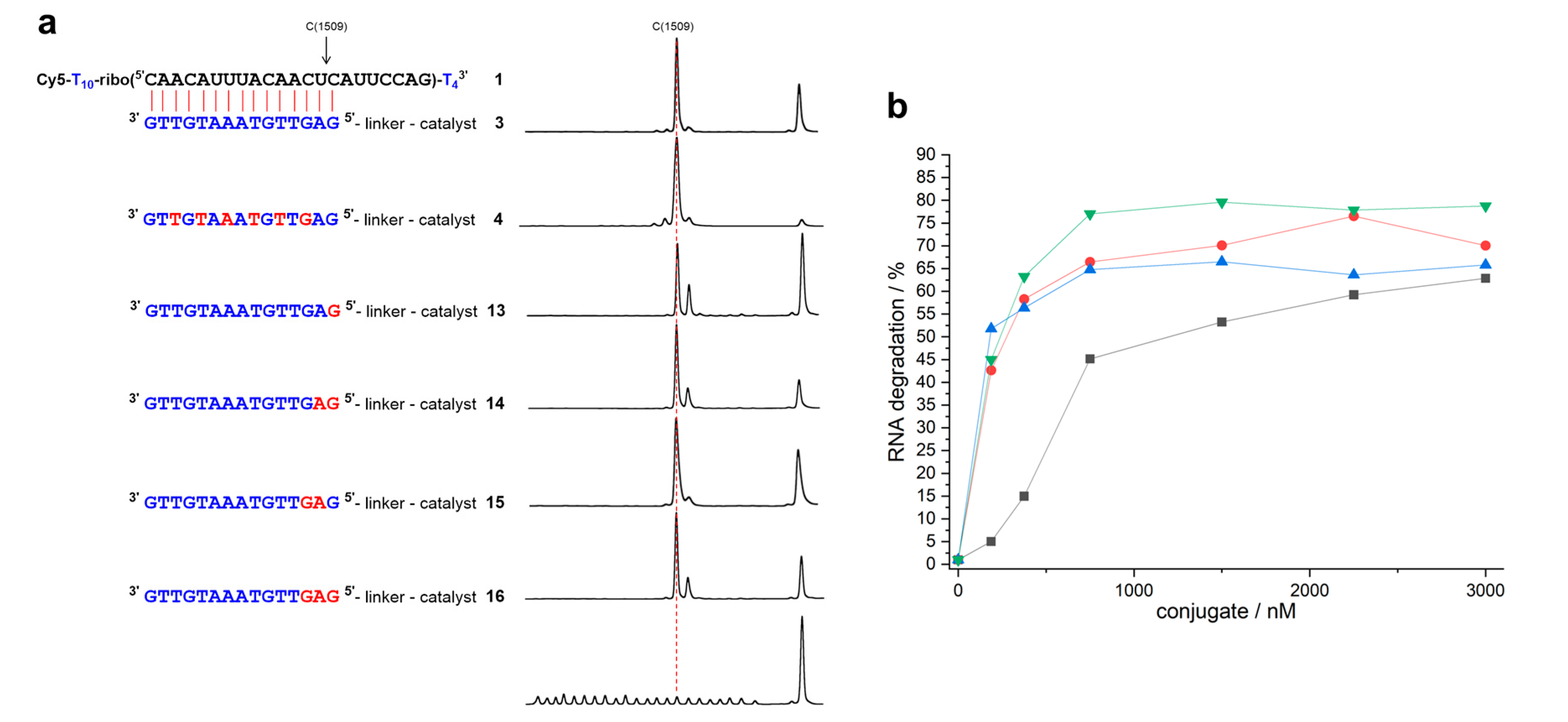
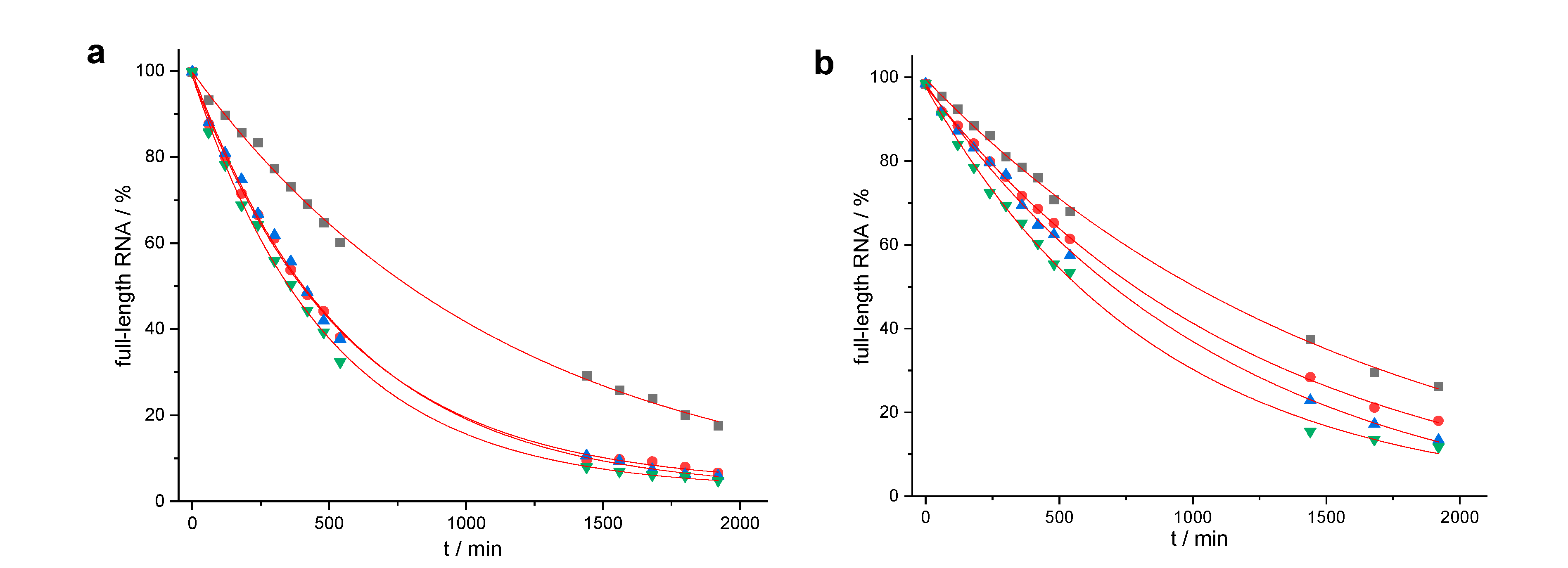
| Conjugate | t1/2 | n − 2 | n − 1 | n | n + 1 | n + 2 | n + 3 |
|---|---|---|---|---|---|---|---|
| 31,3 | 14 ± 0.9 h | 1.4% | 2.5% | 89.0% | 7.1% | ||
| 41,3 | 3.5 ± 0.4 h | 1.6% | 5.8% | 83.9% | 8.6% | ||
| 131 | 13.3 ± 0.8 h | 1.5% | 65.2% | 30.2% | 3.1% | ||
| 141 | 6.4 ± 0.2 h | 1.2% | 75.3% | 22.1% | 1.4% | ||
| 151 | 6.5 ± 0.5 h | 1.5% | 88.3% | 10.1% | |||
| 161 | 5.6 ± 0.1 h | 1.2% | 74.9% | 22.1% | 1.7% | ||
| 52,3 | 15.0 ± 1.7 h | 29.0% | 22.7% | 35.8% | 5.1% | 3.6% | |
| 62,3 | 10.0 ± 0.3 h | 6.4% | 35.0% | 45.9% | 6.9% | 4.5% | 1.3% |
| 172 | 18.3 ± 1.2 h | 5.2% | 4.1% | 52.1% | 22.5% | 9.6% | 2.3% |
| 182 | 14.2 ± 0.7 h | 2.4% | 67.4% | 21.8% | 6.1% | 2.2% | |
| 192 | 13.0 ± 1.5 h | 10.7% | 71.3% | 9.8% | 6.4% | 1.8% | |
| 202 | 9.8 ± 1.0 h | 3.2% | 72.4% | 17.2% | 3.4% | 1.1% |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zellmann, F.; Göbel, M.W. A Trisbenzimidazole Phosphoramidite Building Block Enables High-Yielding Syntheses of RNA-Cleaving Oligonucleotide Conjugates. Molecules 2020, 25, 1842. https://doi.org/10.3390/molecules25081842
Zellmann F, Göbel MW. A Trisbenzimidazole Phosphoramidite Building Block Enables High-Yielding Syntheses of RNA-Cleaving Oligonucleotide Conjugates. Molecules. 2020; 25(8):1842. https://doi.org/10.3390/molecules25081842
Chicago/Turabian StyleZellmann, Felix, and Michael W. Göbel. 2020. "A Trisbenzimidazole Phosphoramidite Building Block Enables High-Yielding Syntheses of RNA-Cleaving Oligonucleotide Conjugates" Molecules 25, no. 8: 1842. https://doi.org/10.3390/molecules25081842