Spectroscopic Study on Pseudomonas Aeruginosa Biofilm in the Presence of the Aptamer-DNA Scaffolded Silver Nanoclusters
Abstract
:1. Introduction
2. Results
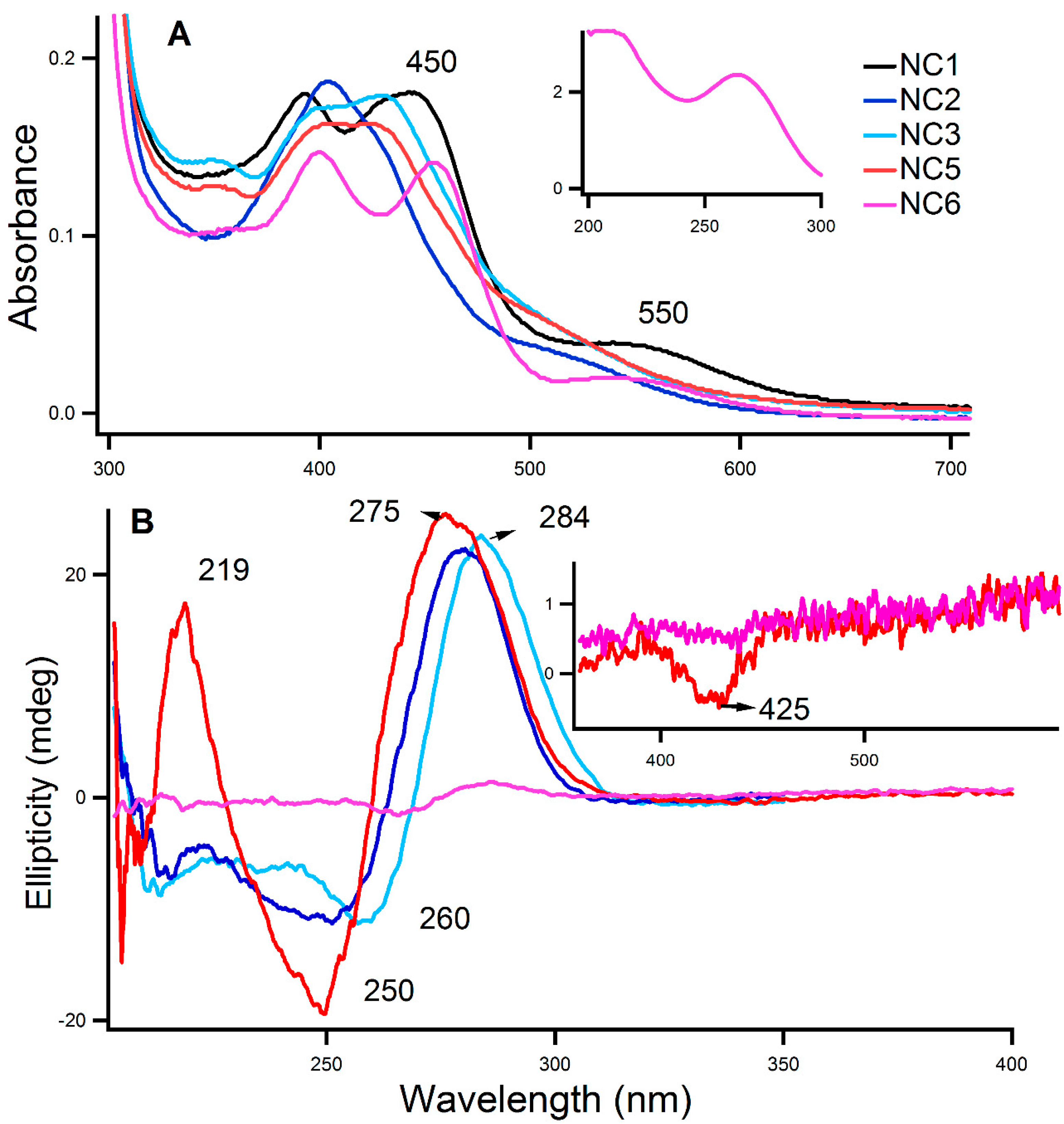
2.1. Characterization of the Aptamer-Templated Ag-NC
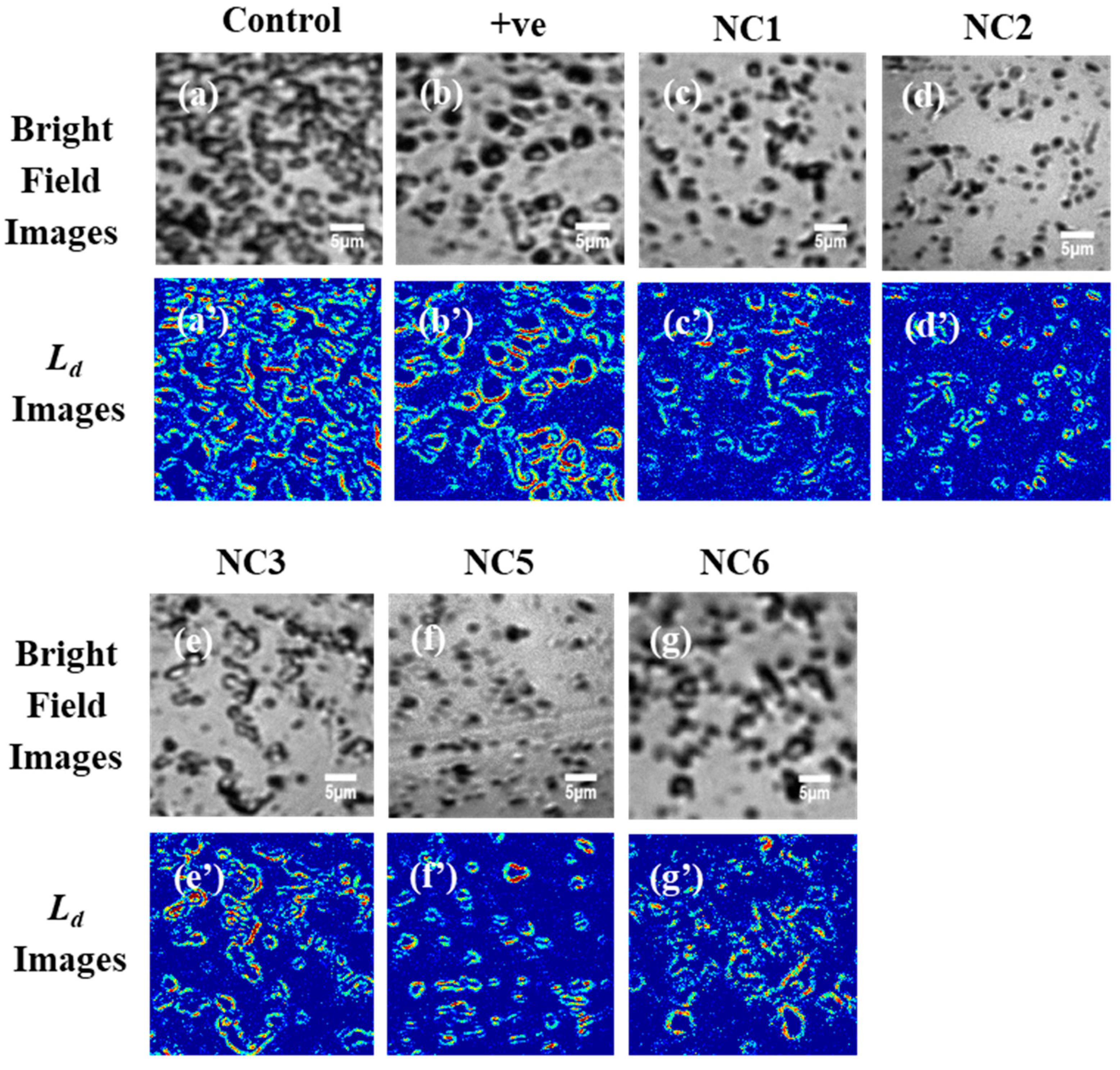
2.2. Influence of the Aptamer-Enclosed Silver Nanoclusters on PA Biofilm
3. Discussions
4. Materials and Methods
4.1. Choosing the Aptamer Sequences
4.2. Preparation of DNA-Templated Silver Nanocluster
4.3. Preparation of Bacterial Samples for Biofilm Study
4.4. Techniques Used to Characterize the Ag-NC and PA Control/Treated with Ag-NC
4.4.1. Steady State Absorption, Fluorescence, and Circular Dichroism
4.4.2. Partial Wave Spectroscopy and the Quantification of Structural Disorder Strength (Ld)
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
References
- Ryder, C.; Byrd, M.; Wozniak, D.J. Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr. Opin. Microbiol. 2007, 10, 644–648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banin, E.; Vasil, M.L.; Greenberg, E.P. Iron and Pseudomonas aeruginosa biofilm formation. Proc. Natl. Acad. Sci. USA 2005, 102, 11076–11081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murga, R.; Miller, J.M.; Donlan, R.M. Biofilm Formation by Gram-Negative Bacteria on Central Venous Catheter Connectors: Effect of Conditioning Films in a Laboratory Model. J. Clin. Microbiol. 2001, 39, 2294–2297. [Google Scholar] [CrossRef] [Green Version]
- Matsukawa, M.; Greenberg, E.P. Putative Exopolysaccharide Synthesis Genes Influence Pseudomonas aeruginosa Biofilm Development. J. Bacteriol. 2004, 186, 4449–4456. [Google Scholar] [CrossRef] [Green Version]
- Colvin, K.M.; Alnabelseya, N.; Baker, P.; Whitney, J.C.; Howell, P.L.; Parsek, M.R. PelA Deacetylase Activity Is Required for Pel Polysaccharide Synthesis in Pseudomonas aeruginosa. J. Bacteriol. 2013, 195, 2329–2339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Parsek, M.R.; Wozniak, D.J.; Ma, L.Z. A spider web strategy of type IV pili-mediated migration to build a fibre-like Psl polysaccharide matrix in Pseudomonas aeruginosa biofilms. Environ. Microbiol. 2013, 15, 2238–2253. [Google Scholar] [CrossRef] [Green Version]
- Zhao, K.; Tseng, B.S.; Beckerman, B.; Jin, F.; Gibiansky, M.L.; Harrison, J.J.; Luijten, E.; Parsek, M.R.; Wong, G.C. Psl trails guide exploration and microcolony formation in Pseudomonas aeruginosa biofilms. Nature 2013, 497, 388–391. [Google Scholar] [CrossRef] [Green Version]
- Monte, J.; Abreu, A.C.; Borges, A.; Simões, L.C.; Simões, M. Antimicrobial Activity of Selected Phytochemicals against Escherichia coli and Staphylococcus aureus and Their Biofilms. Pathogens 2014, 3, 473–498. [Google Scholar] [CrossRef] [Green Version]
- Auger, S.; RamaRao, N.; Faille, C.; Fouet, A.; Aymerich, S.; Gohar, M. Biofilm Formation and Cell Surface Properties among Pathogenic and Nonpathogenic Strains of the Bacillus cereus Group. Appl. Environ. Microbiol. 2009, 75, 6616–6618. [Google Scholar] [CrossRef] [Green Version]
- Weigert, C. Zur Technik der mikroskopischen Bakterienuntersuchungen. Arch. Mikrosk. Anat. 1878, 15, 258–260. [Google Scholar] [CrossRef] [Green Version]
- Pires, D.P.; Cerqueira, M.A.; Faustino, A.; Azeredo, J. Use of newly isolated phages for control of Pseudomonas aeruginosa PAO1 and ATCC 10145 biofilms. Res. Microbiol. 2011, 162, 798–806. [Google Scholar] [CrossRef] [Green Version]
- Rybtke, M.; Hultqvist, L.D.; Givskov, M.; Tolker-Nielsen, T. Pseudomonas aeruginosa Biofilm Infections: Community Structure, Antimicrobial Tolerance and Immune Response. J. Mol. Boil. 2015, 427, 3628–3645. [Google Scholar] [CrossRef] [PubMed]
- Mah, T.-F.; Pitts, B.; Pellock, B.; Walker, G.C.; Stewart, P.S.; O’Toole, G.A. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 2003, 426, 306–310. [Google Scholar] [CrossRef] [PubMed]
- Ramasamy, M.; Lee, J. Recent Nanotechnology Approaches for Prevention and Treatment of Biofilm-Associated Infections on Medical Devices. BioMed Res. Int. 2016, 2016, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, W.; Jiang, C.J.C.; Roy, V.A.L. Recent progress in magnetic iron oxide-semiconductor composite nanomaterials as promising photocatalysts. Nanoscale 2015, 7, 38–58. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Shi, L.; Su, L.; Ren, Q.; Jutte, P.C.; Ren, Y.; Busscher, H.J. Nanotechnology-based antimicrobials and delivery systems for biofilm-infection control. Chem. Soc. Rev. 2019, 48, 428–446. [Google Scholar] [CrossRef]
- Wasukan, N.; Srisun, S.; Kornphimol, K.; Boonrungsiman, S.; Maniratanachote, R. Determination of silver in personal care nanoproductsand effects on dermal exposure. J. Nanopart. Res. 2015, 17, 425–434. [Google Scholar] [CrossRef]
- Prokopovich, P.; Köbrick, M.; Brousseau, E.; Perni, S. Potent antimicrobial activity of bone cement encapsulating silver nanoparticles capped with oleic acid. J. Biomed. Mater. Res. Part B Appl. Biomater. 2014, 103, 273–281. [Google Scholar] [CrossRef] [Green Version]
- Gurunathan, S.; Han, J.W.; Kwon, D.-N.; Kim, J.-H. Enhanced antibacterial and anti-biofilm activities of silver nanoparticles against Gram-negative and Gram-positive bacteria. Nanoscale Res. Lett. 2014, 9, 373. [Google Scholar] [CrossRef] [Green Version]
- Sengupta, B.; Sinha, S.S.; Garner, B.L.; Arany, I.; Corley, C.; Cobb, K.; Brown, E.; Ray, P.C. Influence of Aptamer-Enclosed Silver Nanocluster on the Prevention of Biofilm by Bacillus thuringiensis. Nanosci. Nanotechnol. Lett. 2016, 8, 1054–1060. [Google Scholar] [CrossRef]
- Wu, J.; Li, F.; Hu, X.; Lu, J.; Sun, X.; Gao, J.-Q.; Ling, D. Responsive Assembly of Silver Nanoclusters with a Biofilm Locally Amplified Bactericidal Effect to Enhance Treatments against Multi-Drug-Resistant Bacterial Infections. ACS Central Sci. 2019, 5, 1366–1376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soundy, J.; Day, D.J. Selection of DNA aptamers specific for live Pseudomonas aeruginosa. PLoS ONE 2017, 12, e0185385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, R.; Dhiman, A.; Kapil, A.; Bansal, V.; Sharma, T.K. Aptamer-mediated colorimetric and electrochemical detection of Pseudomonas aeruginosa utilizing peroxidase-mimic activity of gold NanoZyme. Anal. Bioanal. Chem. 2019, 411, 1229–1238. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Setyawati, M.I.; Tan, A.S.; Ong, C.; Leong, D.T.; Xie, J. Highly luminescent silver nanoclusters with tunable emissions: Cyclic reduction-decomposition synthesis and antimicrobial properties. NPG Asia Mater. 2013, 5, e39. [Google Scholar] [CrossRef]
- Soundy, J.; Day, D.J. Delivery of antibacterial silver nanoclusters to Pseudomonas aeruginosa using species-specific DNA aptamers. J. Med Microbiol. 2020, 69, 640–652. [Google Scholar] [CrossRef] [PubMed]
- Almabadi, H.M.; Nagesh, P.K.; Sahay, P.; Bhandari, S.; Eckstein, E.C.; Jaggi, M.; Chauhan, S.C.; Yallapu, M.M.; Pradhan, P.; Yallappu, M.M. Optical study of chemotherapy efficiency in cancer treatment via intracellular structural disorder analysis using partial wave spectroscopy. J. Biophotonics 2018, 11, e201800056. [Google Scholar] [CrossRef] [Green Version]
- Adhikari, P.; Nagesh, P.K.B.; Alharthi, F.; Chauhan, S.C.; Jaggi, M.; Yallapu, M.M.; Pradhan, P. Optical detection of the structural properties of tumor tissue generated by xenografting of drug-sensitive and drug-resistant cancer cells using partial wave spectroscopy (PWS). Biomed. Opt. Express 2019, 10, 6422–6431. [Google Scholar] [CrossRef]
- Ritchie, C.M.; Johnsen, K.R.; Kiser, J.R.; Antoku, Y.; Dickson, R.M.; Petty, J.T. Ag Nanocluster Formation Using a Cytosine Oligonucleotide Template. J. Phys. Chem. C 2007, 111, 175–181. [Google Scholar] [CrossRef] [Green Version]
- Sengupta, B.; Reilly, S.M.; Davis, D.E.; Harris, K.; Wadkins, R.M.; Ward, D.; Gholar, D.; Hampton, C. Excited State Proton Transfer of Natural Flavonoids and Their Chromophores in Duplex and Tetraplex DNAs. J. Phys. Chem. B 2014, 119, 2546–2556. [Google Scholar] [CrossRef] [Green Version]
- Sengupta, B.; Springer, K.; Buckman, J.G.; Story, S.P.; Abe, O.H.; Hasan, W.Z.W.; Prudowsky, Z.D.; Rudisill, S.E.; Degtyareva, N.N.; Petty, J.T. DNA Templates for Fluorescent Silver Clusters and I-Motif Folding. J. Phys. Chem. C 2009, 113, 19518–19524. [Google Scholar] [CrossRef]
- Sengupta, B.; Corley, C.; Cobb, K.; Saracino, A.; Jockusch, S. DNA Scaffolded Silver Clusters: A Critical Study. Molecules 2016, 21, 216. [Google Scholar] [CrossRef] [Green Version]
- Petty, J.T.; Fan, C.; Story, S.P.; Sengupta, B.; Iyer, A.S.J.; Prudowsky, Z.; Dickson, R.M. DNA Encapsulation of 10 Silver Atoms Producing a Bright, Modulatable, Near-Infrared-Emitting Cluster. J. Phys. Chem. Lett. 2010, 1, 2524–2529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subramanian, H.; Pradhan, P.; Liu, Y.; Capoglu, I.R.; Rogers, J.D.; Roy, H.K.; Brand, R.E.; Backman, V. Partial wave microscopic spectroscopy detects sub-wavelength refractive index fluctuations: An application to cancer diagnosis. Opt. Lett. 2009, 34, 518–520. [Google Scholar] [CrossRef]
- Subramanian, H.; Roy, H.K.; Pradhan, P.; Goldberg, M.J.; Muldoon, J.; Brand, R.E.; Sturgis, C.D.; Hensing, T.; Ray, D.; Bogojevic, A.; et al. Nanoscale cellular changes in field carcinogenesis detected by partial wave spectroscopy. Cancer Res. 2009, 69, 5357–5363. [Google Scholar] [CrossRef] [Green Version]
- Subramanian, H.; Pradhan, P.; Liu, Y.; Capoglu, I.R.; Li, X.; Rogers, J.D.; Heifetz, A.; Kunte, D.; Roy, H.K.; Taflove, A.; et al. Optical methodology for detecting histologically unapparent nanoscale consequences of genetic alterations in biological cells. Proc. Natl. Acad. Sci. USA 2008, 105, 20118–20123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhandari, S.; Shukla, P.K.; Almabadi, H.M.; Sahay, P.; Rao, R.; Pradhan, P. Optical study of stress hormone-induced nanoscale structural alteration in brain using partial wave spectroscopic microscopy. J. Biophotonics 2019, 12, 201800002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bos, R.; Ren, Q.; Busscher, H.J. Physico-chemistry of initial microbial adhesive interactions--its mechanisms and methods for study. FEMS Microbiol. Rev. 1999, 23, 179–230. [Google Scholar] [CrossRef]
- Lemos, M.; Gomes, I.; Mergulhão, F.; Melo, L.; Simões, M. The effects of surface type on the removal of Bacillus cereus and Pseudomonas fluorescens single and dual species biofilms. Food Bioprod. Process. 2015, 93, 234–241. [Google Scholar] [CrossRef] [Green Version]
- Geoghegan, M.; Andrews, J.S.; Biggs, C.A.; Eboigbodin, K.E.; Elliott, D.R.; Rolfe, S.A.; Scholes, J.D.; Ojeda, J.; Romero-González, M.; Edyvean, R.G.J.; et al. The polymer physics and chemistry of microbial cell attachment and adhesion. Faraday Discuss. 2008, 139, 85–103. [Google Scholar] [CrossRef]
- Carniello, V.; Peterson, B.W.; Ren, Q.; Busscher, H.J. Physico-chemistry from initial bacterial adhesion to surface-programmed biofilm growth. Adv. Colloid Interface Sci. 2018, 261, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Petty, J.T.; Sergev, O.O.; Kantor, A.G.; Rankine, I.J.; Ganguly, M.; David, F.D.; Wheeler, S.K.; Wheeler, J.F. Ten-Atom Silver Cluster Signaling and Tempering DNA Hybridization. Anal. Chem. 2015, 87, 5302–5309. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Castanon, G.-A.; Niño-Martínez, N.; Martínez-Gutierrez, F.; Martínez-Mendoza, J.R.; Ruiz, F. Synthesis and antibacterial activity of silver nanoparticles with different sizes. J. Nanoparticle Res. 2008, 10, 1343–1348. [Google Scholar] [CrossRef]
- Rashid, M.U.; Bhuiyan, K.H.; Quayum, M.E. Synthesis of Silver Nano Particles (Ag-NPs) and their uses for Quantitative Analysis of Vitamin C Tablets. Dhaka Univ. J. Pharm. Sci. 2013, 12, 29–33. [Google Scholar] [CrossRef]
Sample Availability: Samples of the silver nanoclusters on DNA templates can be made available from the authors, if needed. |

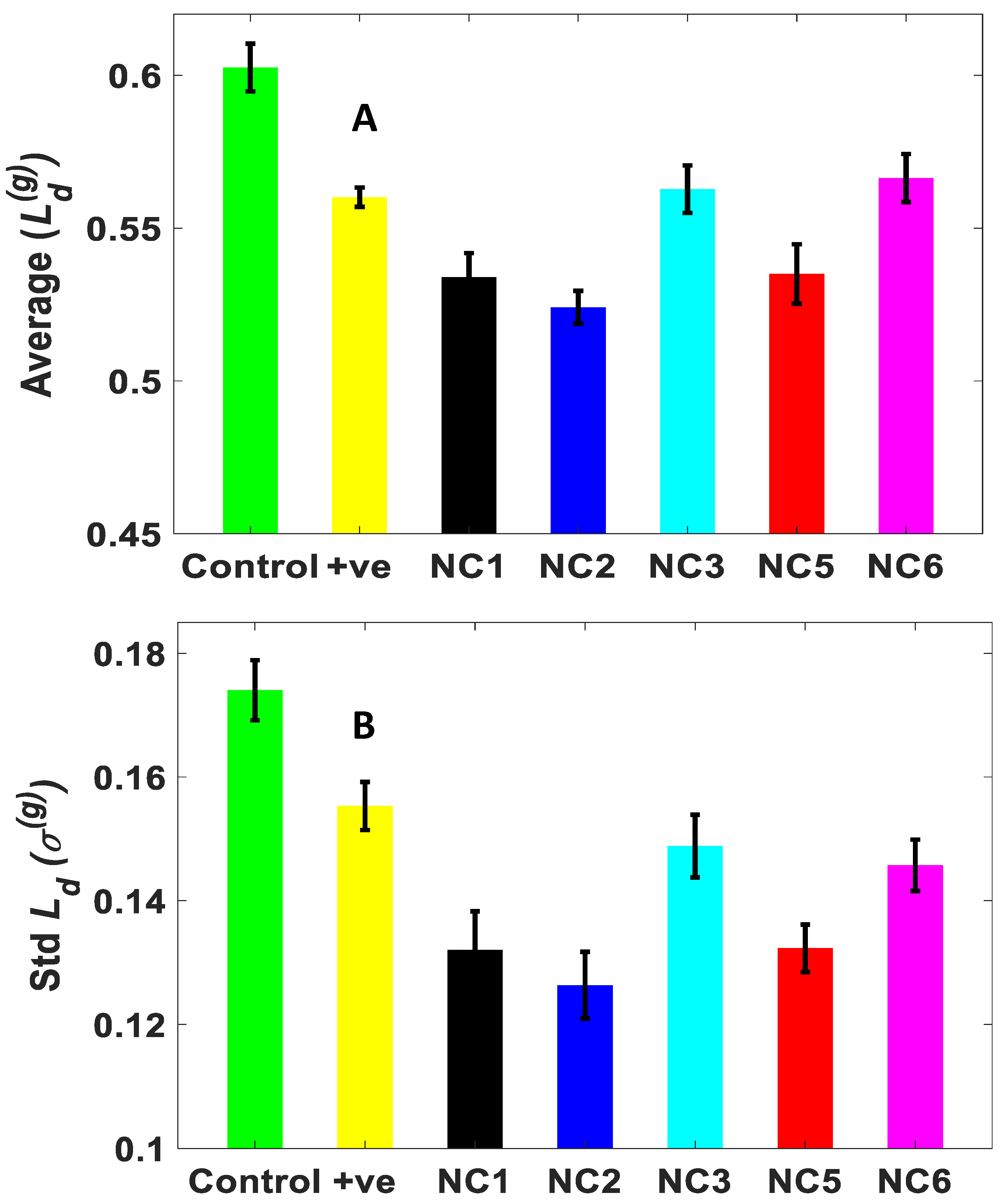
| Samples | ΔLd(g) (%) | Δσ(g) (%) |
|---|---|---|
| Positive control | 7.04068 | 10.7574 |
| NC1 | 11.38513 | 24.1122 |
| NC2 | 13.0183 | 27.3992 |
| NC3 | 6.6087 | 14.4696 |
| NC5 | 11.2092 | 23.9627 |
| NC6 | 6.0012 | 16.2625 |
| DNA Name | Sequence | Absorption Wavelength (nm) with Absorbance (OD) of the Ag-NC |
|---|---|---|
| NC1 | 5′-TAC TTC CGC ACC CTC CTA CA-3′ | 442 (0.18), 550 (sh) |
| NC2 | 5′-CCC CCG TTG CTT TCG CTT TTC CTT TCG CTT TTG TTC GTT TCG TCC CTG CTT CCT TTC TTG-3′ | 430 (0.15), 512 (sh) |
| NC3 | 5′-CCC TTT CCC TTT CCC ATT CCC GTT CCC TTT CCC TTT CCC ATT CCC GTT A-3′ | 431 (0.18), 511 (sh) |
| NC5 | 5′-ATGAGAGCGTCGGTGTGGTA- CCC TTT CCC TTT CCC ATT CCC GTT CCC TTT CCC TTT CCC ATT CCC GTT A -TACTTCCGCACCCTCCTACA-3′ | 427 (0.16), 511 (sh) |
| NC6 | 5′-ATG AGA GCG TCG GTG TGG TA-3′ | 454 (0.14), 560 (0.017) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sengupta, B.; Adhikari, P.; Mallet, E.; Havner, R.; Pradhan, P. Spectroscopic Study on Pseudomonas Aeruginosa Biofilm in the Presence of the Aptamer-DNA Scaffolded Silver Nanoclusters. Molecules 2020, 25, 3631. https://doi.org/10.3390/molecules25163631
Sengupta B, Adhikari P, Mallet E, Havner R, Pradhan P. Spectroscopic Study on Pseudomonas Aeruginosa Biofilm in the Presence of the Aptamer-DNA Scaffolded Silver Nanoclusters. Molecules. 2020; 25(16):3631. https://doi.org/10.3390/molecules25163631
Chicago/Turabian StyleSengupta, Bidisha, Prakash Adhikari, Esther Mallet, Ronald Havner, and Prabhakar Pradhan. 2020. "Spectroscopic Study on Pseudomonas Aeruginosa Biofilm in the Presence of the Aptamer-DNA Scaffolded Silver Nanoclusters" Molecules 25, no. 16: 3631. https://doi.org/10.3390/molecules25163631








