Discovery of Indoleamine 2,3-Dioxygenase 1 (IDO-1) Inhibitors Based on Ortho-Naphthaquinone-Containing Natural Product
Abstract
:1. Introduction
2. Results and Discussion
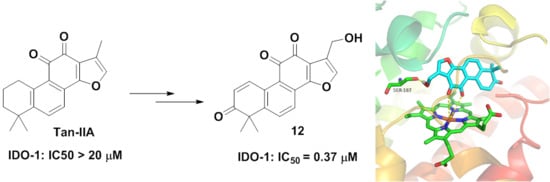
2.1. Chemistry
2.2. Biological Evaluation
2.3. Molecular Docking Study
3. Materials and Methods
3.1. Chemistry
3.1.1. General Chemistry
3.1.2. Procedure for the Preparation of Compounds
Synthesis of 1,6,6-trimethyl-6,7-dihydrophenanthro[1,2-b]furan-10,11-dione (9) [33]
Synthesis of 1,6,6-trimethylphenanthro[1,2-b]furan-7,10,11(6H)-trione (11)
Synthesis of 1-(hydroxymethyl)-6,6-dimethylphenanthro[1,2-b]furan-7,10,11(6H)-trione (12)
Synthesis of 7-hydroxy-1,6,6-trimethyl-6,7-dihydrophenanthro[1,2-b]furan-10,11-dione (10) [33]
Synthesis of 1,6,6-trimethyl-7,8-dihydrophenanthro[1,2-b]furan-9,10,11(6H)-trione (13) and 1,6,6-trimethylphenanthro[1,2-b]furan-9,10,11(6H)-trione (14)
Synthesis of 1,6,6-trimethylphenanthro[1,2-b]furan-9,10,11(6H)-trione (14)
Synthesis of 1-(hydroxymethyl)-6,6-dimethylphenanthro[1,2-b]furan-9,10,11(6H)-trione (15)
3.2. Biology
3.3. Docking Study
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Robert, C.; Schachter, J.; Long, G.V.; Arance, A.; Grob, J.J.; Mortier, L.; Daud, A.; Carlino, M.S.; McNeil, C.; Lotem, M.; et al. Pembrolizumab versus Ipilimumab in Advanced Melanoma. N. Engl. J. Med. 2015, 372, 2521–2532. [Google Scholar] [CrossRef]
- Hodi, F.S.; O’Day, S.J.; McDermott, D.F.; Weber, R.W.; Sosman, J.A.; Haanen, J.B.; Gonzalez, R.; Robert, C.; Schadendorf, D.; Hassel, J.C.; et al. Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med. 2010, 363, 711–723. [Google Scholar] [CrossRef] [PubMed]
- Yue, E.W.; Sparks, R.; Polam, P.; Modi, D.; Douty, B.; Wayland, B.; Glass, B.; Takvorian, A.; Glenn, J.; Zhu, W.; et al. INCB24360 (Epacadostat), a Highly Potent and Selective Indoleamine-2,3-dioxygenase 1 (IDO-1) Inhibitor for Immuno-oncology. ACS Med. Chem. Lett. 2017, 8, 486–491. [Google Scholar] [CrossRef] [PubMed]
- Masanori, S.; Mark, P.R.; Eric, D.C.; John, H.D. Heme-Containing Oxygenases. Chem. Rev. 1996, 96, 2841–2887. [Google Scholar]
- Yamamoto, S.; Hayaishi, O. Tryptophan pyrrolase of rabbit intestine. d- and l-tryptophan-cleaving enzyme or enzymes. J. Biol. Chem. 1967, 242, 5260–5266. [Google Scholar] [PubMed]
- David, H.M.; Zhou, M.; John, T.A.; Igor, B.; Simon, J.C.; Brendan, M.; Corrie, B.; Andrew, L.M. Prevention of Allogeneic Fetal Rejection by Tryptophan Catabolism. Science 1998, 281, 1191–1193. [Google Scholar]
- Hwu, P.; Du, M.X.; Lapointe, R.; Do, M.; Taylor, M.W.; Young, H.A. Indoleamine 2,3-Dioxygenase Production by Human Dendritic Cells Results in the Inhibition of T Cell Proliferation. J. Immunol. 2000, 164, 3596–3599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terness, P.; Bauer, T.M.; Röse, L.; Dufter, C.; Watzlik, A.; Simon, H.; Opelz, G. Inhibition of Allogeneic T Cell Proliferation by Indoleamine 2,3-Dioxygenase–expressing Dendritic Cells. J. Exp. Med. 2002, 196, 447–457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fallarino, F.; Grohmann, U.; You, S.; McGrath, B.C.; Cavener, D.R.; Vacca, C.; Orabona, C.; Bianchi, R.; Belladonna, M.L.; Volpi, C.; et al. The Combined Effects of Tryptophan Starvation and Tryptophan Catabolites Down-Regulate T Cell Receptor-Chain and Induce a Regulatory Phenotype in Naive T Cells. J. Immunol. 2006, 176, 6752–6761. [Google Scholar] [CrossRef]
- Munn, D.H.; Mellor, A.L. Indoleamine 2,3-dioxygenase and tumor-induced tolerance. J. Clin. Investig. 2007, 117, 1147–1154. [Google Scholar] [CrossRef] [Green Version]
- Uyttenhove, C.; Pilotte, L.; Theate, I.; Stroobant, V.; Colau, D.; Parmentier, N.; Boon, T.; Van den Eynde, B.J. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nat. Med. 2003, 9, 1269–1274. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, A.; Nikaido, T.; Ochiai, K.; Takakura, S.; Saito, M.; Aoki, Y.; Ishii, N.; Yanaihara, N.; Yamada, K.; Takikawa, O.; et al. Indoleamine 2,3-dioxygenase serves as a marker of poor prognosis in gene expression profiles of serous ovarian cancer cells. Clin. Cancer Res. 2005, 11, 6030–6039. [Google Scholar] [CrossRef] [PubMed]
- Hou, D.Y.; Muller, A.J.; Sharma, M.D.; DuHadaway, J.; Banerjee, T.; Johnson, M.; Mellor, A.L.; Prendergast, G.C.; Munn, D.H. Inhibition of indoleamine 2,3-dioxygenase in dendritic cells by stereoisomers of 1-methyl-tryptophan correlates with antitumor responses. Cancer Res. 2007, 67, 792–801. [Google Scholar] [CrossRef] [PubMed]
- Muller, A.J.; DuHadaway, J.B.; Donover, P.S.; Sutanto-Ward, E.; Prendergast, G.C. Inhibition of indoleamine 2,3-dioxygenase, an immunoregulatory target of the cancer suppression gene Bin1, potentiates cancer chemotherapy. Nat. Med. 2005, 11, 312–319. [Google Scholar] [CrossRef] [PubMed]
- Prendergast, G.C.; Malachowski, W.P.; DuHadaway, J.B.; Muller, A.J. Discovery of IDO1 Inhibitors: From Bench to Bedside. Cancer Res. 2017, 77, 6795–6811. [Google Scholar] [CrossRef]
- Sharma, M.D.; Hou, D.Y.; Liu, Y.; Koni, P.A.; Metz, R.; Chandler, P.; Mellor, A.L.; He, Y.; Munn, D.H. Indoleamine 2,3-dioxygenase controls conversion of Foxp3+ Tregs to TH17-like cells in tumor-draining lymph nodes. Blood. 2009, 113, 6102–6111. [Google Scholar] [CrossRef] [Green Version]
- Rohrig, U.F.; Majjigapu, S.R.; Vogel, P.; Zoete, V.; Michielin, O. Challenges in the Discovery of Indoleamine 2,3-Dioxygenase 1 (IDO-1) Inhibitors. J. Med. Chem. 2015, 58, 9421–9437. [Google Scholar] [CrossRef]
- Nakashima, H.; Uto, Y.; Nakata, E.; Nagasawa, H.; Ikkyu, K.; Hiraoka, N.; Nakashima, K.; Sasaki, Y.; Sugimoto, H.; Shiro, Y.; et al. Synthesis and biological activity of 1-methyl-tryptophan-tirapazamine hybrids as hypoxia-targeting indoleamine 2,3-dioxygenase inhibitors. Bioorg. Med. Chem. 2008, 16, 8661–8669. [Google Scholar] [CrossRef]
- Mautino, M.; Kumar, S.; Waldo, J.; Jaipuri, F.; Kesharwani, T. Fused Imidazole Derivatives Useful as IDO Inhibitors. U.S. Patent WO2012/142237, 18 October 2012. [Google Scholar]
- Nakao, M.; Fukushima, T. On the chemical composition of Salvia miltiorrhiza (Chinese drug Tan-shen). J. Pharm. Soc. Jpn. 1934, 54, 154–162. [Google Scholar] [CrossRef]
- Zhou, L.; Zuo, Z.; Chow, M.S. Danshen: An overview of its chemistry, pharmacology, pharmacokinetics, and clinical use. J. Clin. Pharmacol. 2005, 45, 1345–1359. [Google Scholar] [CrossRef]
- Wang, X.H.; Morris-Natschke, S.L.; Lee, K.H. New developments in the chemistry and biology of the bioactive constituents of Tanshen. Med. Res. Rev. 2007, 27, 133–148. [Google Scholar] [CrossRef]
- Dong, Y.; Morris-Natschke, S.L.; Lee, K.H. Biosynthesis, total syntheses, and antitumor activity of tanshinones and their analogs as potential therapeutic agents. Nat. Prod. Rep. 2011, 28, 529–542. [Google Scholar] [CrossRef]
- Don, M.J.; Shen, C.C.; Syu, W.J.; Ding, Y.H.; Sun, C.M. Cytotoxic and aromatic constituents from Salvia miltiorrhiza. Phytochemistry 2006, 67, 497–503. [Google Scholar] [CrossRef]
- Jiao, M.; Ding, C.; Zhang, A. Facile construction of 3-hydroxyphenanthrene-1,4-diones Using a tandem three-step reaction sequence as key intermediates to tanshinone I and its 4-demethylated analogues. Tetrahedron 2014, 70, 2976–2981. [Google Scholar] [CrossRef]
- Jiao, M.; Ding, C.; Zhang, A. Preparation of 2-aryl derivatives of tanshinone I through a palladium-catalyzed Csp2–H activation/arylation approach. Tetrahedron Lett. 2015, 56, 2799–2802. [Google Scholar] [CrossRef]
- Ding, C.; Li, J.; Jiao, M.; Zhang, A. Direct Catalyst-Free C(sp3)-H Acyloxylation of Tanshinone IIA: Regioselective Synthesis of C1-Acyloxyl Derivatives. J. Nat. Prod. 2016, 79, 2514–2520. [Google Scholar] [CrossRef]
- Liang, B.; Yu, S.; Li, J.; Wang, F.; Liang, G.; Zhang, A.; Ding, C. Site-Selective Csp3-H Aryloxylation of Natural Product Tanshinone IIA and Its Analogues. Tetrahedron Lett. 2017, 58, 1822–1825. [Google Scholar] [CrossRef]
- Li, J.; Xue, Y.; Fan, Z.; Ding, C.; Zhang, A. Dihydroxydifluorination of Tanshinones Analogues. J. Org. Chem. 2017, 82, 7388–7393. [Google Scholar] [CrossRef] [PubMed]
- Ding, C.; Tian, Q.; Li, J.; Jiao, M.; Song, S.; Wang, Y.; Miao, Z.-H.; Zhang, A. Structural Modification of Natural Product Tanshinone I Leading to Discovery of Novel Nitrogen-Enriched Derivatives with Enhanced Anticancer Profile and Improved Drug-Like Properties. J. Med. Chem. 2018, 61, 760–776. [Google Scholar] [CrossRef]
- Wang, F.; Yang, H.; Yu, S.; Xue, Y.; Fan, Z.; Liang, G.; Geng, M.; Zhang, A.; Ding, C. Divergent Total Synthesis of (±)-Tanshinol B, (±)-Tanshindiol B, (±)-Tanshindiol C. and Tanshinone I. Org. Biomol. Chem. 2018, 16, 3376–3381. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, W.; Liu, Z.; Ju, Y.; Xu, M.; Zhang, Y.; Wu, X.; Gu, Q.; Wang, Z.; Xu, J. Discovery of indoleamine 2,3-dioxygenase inhibitors using machine learning based virtual screening. Med. Chem. Commun. 2018, 9, 937–945. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Wang, J.; Huang, Z. Tanshinone IIA Derivative Useful in Treatment of Cancer and Its Preparation. Patent CN 105,884,856, 24 August 2016. [Google Scholar]
- Qin, Y.; Su, M.; Jin, Q.; Chen, T.; Wu, X.; Mao, L. 1-Carbonyl Tanshinone IIA Sodium Sulfonate Analogue, Preparation Method and Application. Patent CN 104,910,250, 16 September 2015. [Google Scholar]
- Zhao, Q.; Leng, Y.; Deng, X.; Zhao, Y.; Shen, Y.; Luo, X. Tanshinone Derivative, Its Pharmaceutical Composition, and Medical Application for Treating Diabetes Mellitus and Related Metabolic Diseases. Patent CN 102,603,861, 25 July 2012. [Google Scholar]
- Yu, C.J.; Zheng, M.F.; Kuang, C.X.; Huang, W.D.; Yang, Q. Oren-gedoku-to and its constituents with therapeutic potential in Alzheimer’s disease inhibit indoleamine 2,3-dioxygenase activity in vitro. J. Alzheimers Dis. 2010, 22, 257–266. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds 11, 12, 14 are available from the authors. |



| Compound Number | R1 | R2 | Compound Number | R1 | R2 |
|---|---|---|---|---|---|
| 7a |  | H | 7h |  | H |
| 7b |  | H | 7i |  | H |
| 7c |  |  | 7j |  | H |
| 7d |  |  | 7k |  | H |
| 7e |  | H | 7l |  | H |
| 7f |  | H | 7m |  | H |
| 7g |  | H |
| Compound Number | R1 | R2 | Enzymatic IC50 (μM) or Inhibitory Rate at 20 µM a | Cellular IC50 (μM) a | |
|---|---|---|---|---|---|
 | 6 | H | H | 23.1 ± 1.6% | — b |
| 7a |  | H | 3.72 ± 0.45 | — | |
| 7b |  | H | 4.72 ± 0.30 | — | |
| 7c |  |  | 29.3 ± 2.2% | — | |
| 7d |  |  | 21.5 ± 1.1% | — | |
| 7e |  | H | 7.47 ± 3.58 | — | |
| 7f |  | H | 2.74 ± 0.10 | — | |
| 7g |  | H | 7.05 ± 2.46 | — | |
| 7h |  | H | 48.0 ± 3.2% | — | |
| 7i |  | H | 54.5 ± 2.3% | — | |
| 7j |  | H | 60.4 ± 3.9% | — | |
| 7k |  | H | 4.60 ± 2.81 | — | |
| 7l |  | H | 5.71 ± 0.47 | — | |
| 7m |  | H | 4.96 ± 2.02 | — | |
| 8 | OH | H | 2.83 ± 0.12 | — | |
 | 9 | H | 61.2 ± 2.1% | >10 | |
| 10 | OH | 3.10 ± 1.02 | >10 | ||
 | 11 | H | 0.84 ± 0.26 | >10 | |
| 12 | OH | 0.37 ± 0.02 | 3.85 ± 0.86 | ||
 | 14 | H | 52.7 ± 2.5% | >10 | |
| 15 | OH | 68.0 ± 8.6% | — | ||
| Epacadostat | 0.086 ± 0.009 | 0.023 ± 0.003 | |||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, H.; Sun, P.; Guo, W.; Wang, Y.; Zhang, A.; Meng, L.; Ding, C. Discovery of Indoleamine 2,3-Dioxygenase 1 (IDO-1) Inhibitors Based on Ortho-Naphthaquinone-Containing Natural Product. Molecules 2019, 24, 1059. https://doi.org/10.3390/molecules24061059
Zhao H, Sun P, Guo W, Wang Y, Zhang A, Meng L, Ding C. Discovery of Indoleamine 2,3-Dioxygenase 1 (IDO-1) Inhibitors Based on Ortho-Naphthaquinone-Containing Natural Product. Molecules. 2019; 24(6):1059. https://doi.org/10.3390/molecules24061059
Chicago/Turabian StyleZhao, Hongchuan, Pu Sun, Wei Guo, Yi Wang, Ao Zhang, Linghua Meng, and Chunyong Ding. 2019. "Discovery of Indoleamine 2,3-Dioxygenase 1 (IDO-1) Inhibitors Based on Ortho-Naphthaquinone-Containing Natural Product" Molecules 24, no. 6: 1059. https://doi.org/10.3390/molecules24061059