A Study on Tissue-Specific Metabolite Variations in Polygonum cuspidatum by High-Resolution Mass Spectrometry-Based Metabolic Profiling
Abstract
:1. Introduction
2. Results and Discussion
2.1. Optimization of the Extraction Procedure and MS Conditions
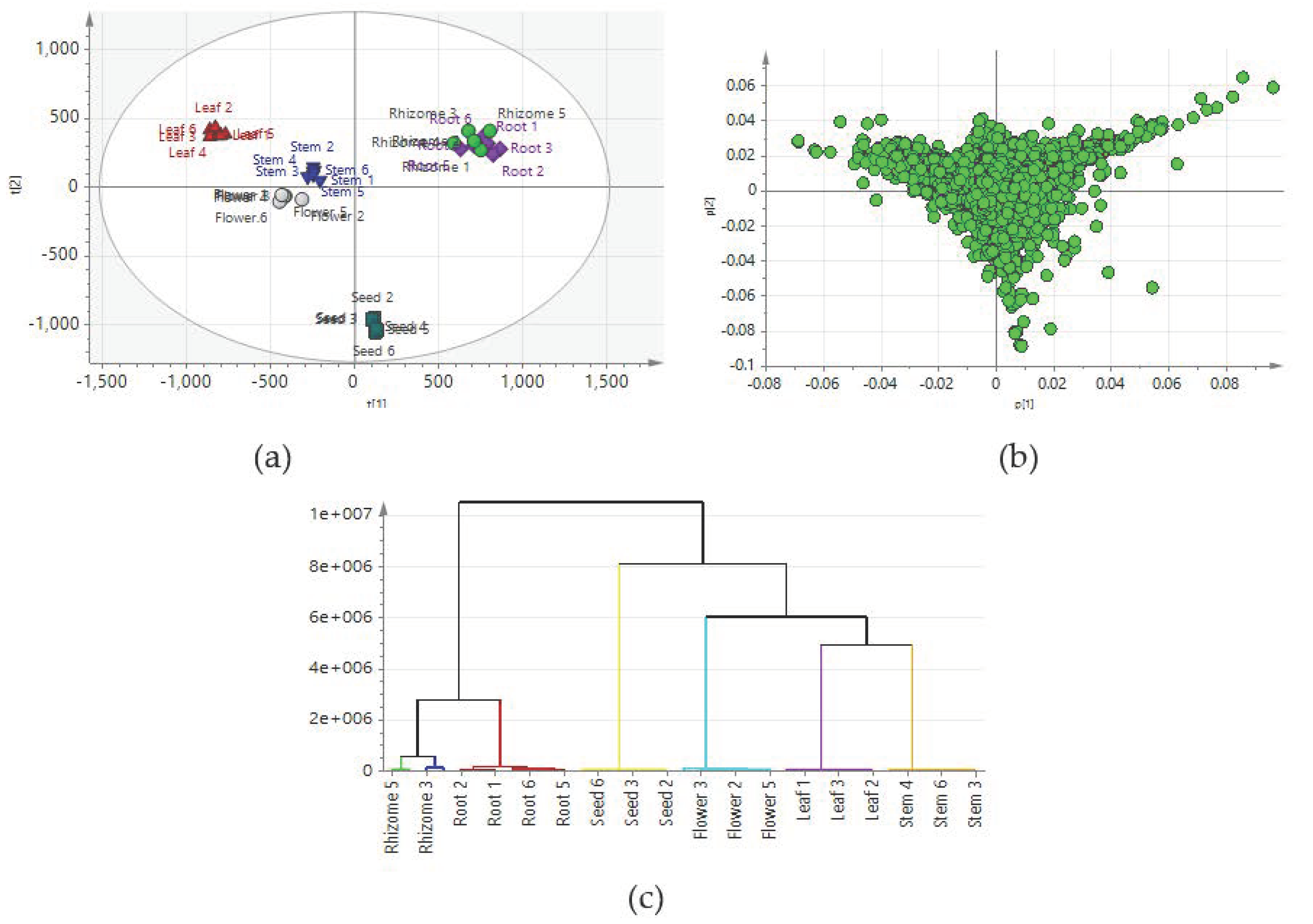
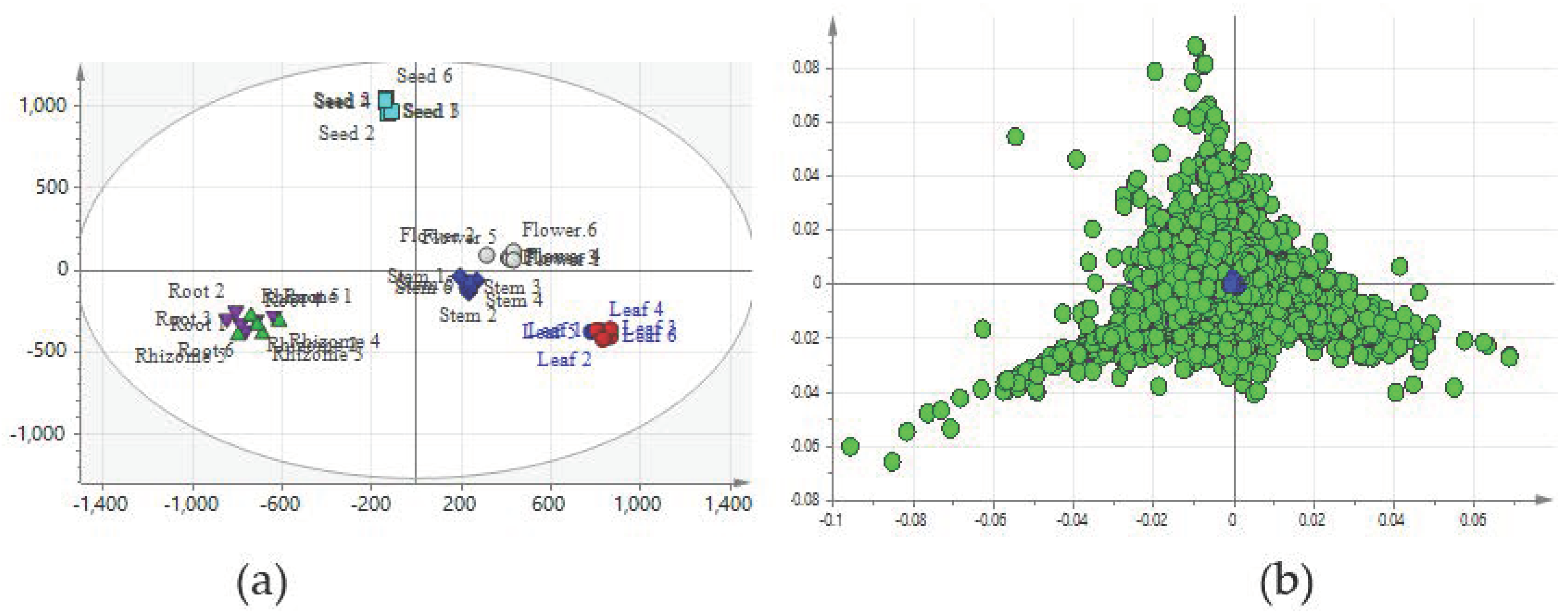
2.2. Comparison of Metabolic Profiles of Different Tissues
2.3. Construction of Predictive Models for the PCA
2.4. Tentative Identification of Bioactive Metabolites in P. cuspidatum
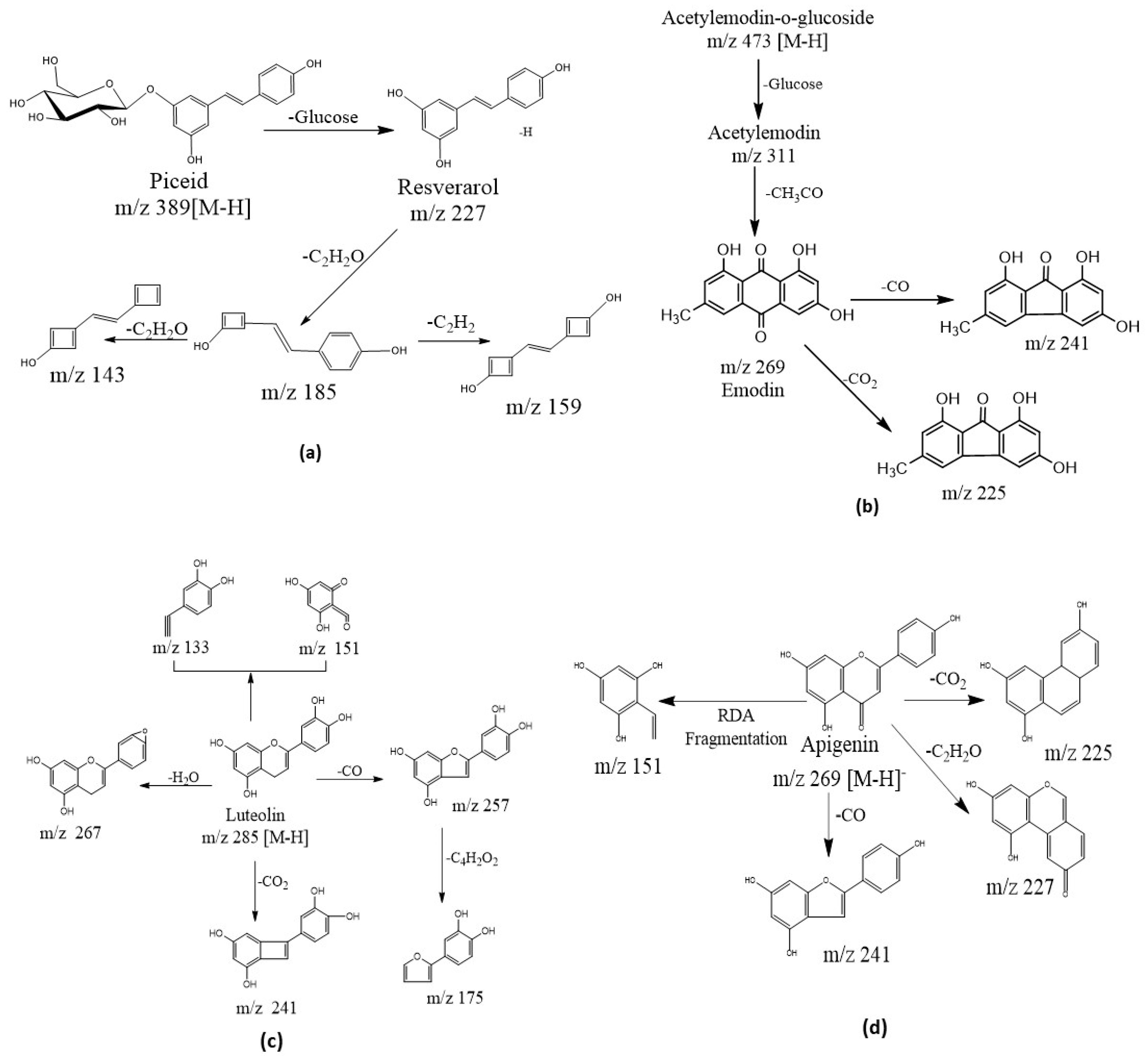
2.4.1. Stilbenes and Their Derivatives
2.4.2. Anthraquinones
2.4.3. Flavonoids, Isoflavonoids and Other Compounds
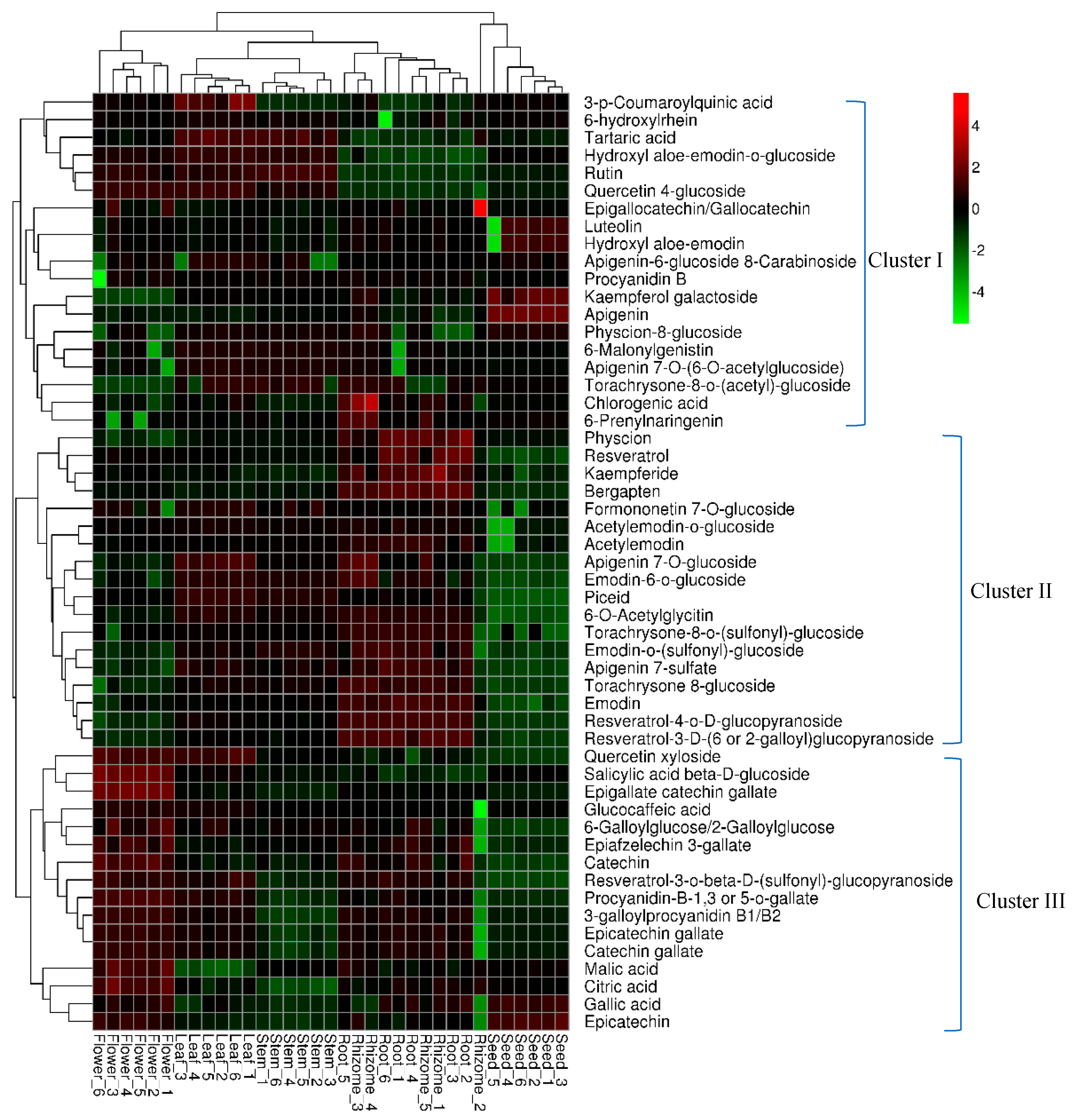
2.5. Global Distribution and Relative Quantitative Analysis of Identified Metabolites
2.6. Identification of Chemical Markers
3. Materials and Methods
3.1. Plant Materials and Chemicals
3.2. Metabolite Extractions
3.3. LC-MS Analysis
3.4. Metabolomics Profiles and MS Data Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Peng, W.; Qin, R.; Li, X.; Zhou, H. Botany, phytochemistry, pharmacology, and potential application of Polygonum cuspidatum Sieb.et Zucc.: A review. J. Ethnopharmacol. 2013, 148, 729–745. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Wang, M.; Guo, H.; Tian, Y.; Zhang, Z.; Song, R. Profiling of components of rhizoma et radix polygoni cuspidati by high-performance liquid chromatography with ultraviolet diode-array detector and ion trap/time-of-flight mass spectrometric detection. Pharmacogn. Mag. 2015, 11, 486–501. [Google Scholar] [PubMed] [Green Version]
- Zhang, H.; Li, C.; Kwok, S.T.; Zhang, Q.W.; Chan, S.W. A Review of the Pharmacological Effects of the Dried Root of Polygonum cuspidatum (Hu Zhang) and Its Constituents. Evid. Based Complement. Altern. Med. 2013, 2013, 1–13. [Google Scholar] [Green Version]
- Zhang, H.; Guo, Y.W.; Yang, Y.D.; Ma, Y.J.; Liu, B.-Y.; Ye, H.-C.; Wang, H.; Ma, L.-Q. Cloning of a novel type III polyketide synthase encoded by a three-intron gene from Polygonum cuspidatum. J. Plant Biochem. Biotechnol. 2012, 23, 104–111. [Google Scholar] [CrossRef]
- Xu, M.; Jin, Z.; Ohm, J.-B.; Schwarz, P.; Rao, J.; Chen, B. Improvement of the Antioxidative Activity of Soluble Phenolic Compounds in Chickpea by Germination. J. Agric. Food Chem. 2018, 66, 6179–6187. [Google Scholar] [CrossRef] [PubMed]
- Valente, I.M.; Maia, M.R.G.; Malushi, N.; Oliveira, H.M.; Papa, L.; Rodrigues, J.A.; Fonseca, A.J.M.; Cabrita, A.R.J. Profiling of phenolic compounds and antioxidant properties of European varieties and cultivars of Vicia faba L. pods. Phytochemistry 2018, 152, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Jayatilake, G.S.; Jayasuriya, H.; Lee, E.S.; Koonchanok, N.M.; Geahlen, R.L.; Ashendel, C.L.; McLaughlin, J.L.; Chang, C.J. Kinase Inhibitors from Polygonum cuspidatum. J. Nat. Prod. 1993, 56, 1805–1810. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Fu, J.; Yin, X.B.; Cao, S.L.; Li, X.C.; Lin, L.F.; Huyiligeqi Ni, J. Emodin: A Review of its Pharmacology, Toxicity and Pharmacokinetics. Phytother. Res. 2016, 30, 1207–1218. [Google Scholar] [CrossRef]
- Editorial Committee of Chinese Pharmacopoeia. Chinese Pharmacopoeia; Editorial Committee of Chinese Pharmacopoeia: Beijing, China, 2015; Volume 1, p. 208. [Google Scholar]
- Ma, Y. The Resources, Quality and Pharmaceutical Efficacy of Polygonum cuspidatum Sieb. et Zucc.; Chendu Unversity of TCM: Chendu, China, 2006. [Google Scholar]
- Ma, P. Pharmacognostic Studies of Polygonum cuspidatum Sieb. et Zucc. (Polygonaceae); Union Medical College: Beijing, China, 2013. [Google Scholar]
- Lin, S.P.; Chu, P.M.; Tsai, S.Y.; Wu, M.H.; Hou, Y.C. Pharmacokinetics and tissue distribution of resveratrol, emodin and their metabolites after intake of Polygonum cuspidatum in rats. J. Ethnopharmacol. 2012, 144, 671–676. [Google Scholar] [CrossRef]
- Dong, J.; Wang, H.; Wan, L.; Yuki, H.; Chen, S. Identification and determination of major constituents in Polygonum cuspidatum Sieb. et Zucc. by high performance liquid chromatography/electrospray ionization-ion trap-time-of-flight mass spectrometry. Chin. J. Chromatagr. 2009, 27, 425–430. [Google Scholar]
- Kim, Y.S.; Hwang, C.S.; Shin, D.H. Volatile constituents from the leaves of Polygonum cuspidatum S. et Z. and their anti-bacterial activities. Food Microbiol. 2005, 22, 139–144. [Google Scholar] [CrossRef]
- Ding, M.Y.; Jiang, Y.; Yu, X.A.; Zhang, D.; Li, J.; Wang, H.; Shen, J.Y.; Gao, X.M.; Chang, Y.X. Screening of Combinatorial Quality Markers for Natural Products by Metabolomics Coupled With Chemometrics. A Case Study on Pollen Typhae. Front. Pharmacol. 2018, 9, 691. [Google Scholar] [CrossRef] [PubMed]
- Geng, Z.; Liu, Y.F.; Gou, Y.; Zhou, Q.M.; He, C.J.; Guo, L.; Zhou, J.; Xiong, L. Metabolomics Study of Cultivated Bulbus Fritillariae Cirrhosae at Different Growth Stages using UHPLC-QTOF-MS Coupled with Multivariate Data Analysis. Phytochem. Anal. 2018, 29, 290–299. [Google Scholar] [CrossRef] [PubMed]
- Mais, E.; Alolga, R.N.; Wang, S.L.; Linus, L.O.; Yin, X.J.; Qi, L.W. A comparative UPLC-Q/TOF-MS-based metabolomics approach for distinguishing Zingiber officinale Roscoe of two geographical origins. Food Chem. 2018, 240, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, L.; Xiao, Y.-Q.; Yao, J.-Q.; Li, P.-Y.; Yu, D.-R.; Ma, Y.-L. Global metabolite profiling and diagnostic ion filtering strategy by LC–QTOF MS for rapid identification of raw and processed pieces of Rheum palmatum L. Food Chem. 2016, 192, 531–540. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.B.; Lane, M.E.; Yang, M.; Heinrich, M. A Hexa-Herbal TCM Decoction Used to Treat Skin Inflammation: An LC-MS-Based Phytochemical Analysis. Planta Med. 2016, 82, 1134–1141. [Google Scholar] [CrossRef] [Green Version]
- Malongane, F.; McGaw, L.J.; Nyoni, H.; Mudau, F.N. Metabolic profiling of four South African herbal teas using high resolution liquid chromatography-mass spectrometry and nuclear magnetic resonance. Food Chem. 2018, 257, 90–100. [Google Scholar] [CrossRef]
- Wang, H.; Feng, F. Identification of components in Zhi-Zi-Da-Huang decoction by HPLC coupled with electrospray ionization tandem mass spectrometry, photodiode array and fluorescence detectors. J. Pharm. Biomed. Anal. 2009, 49, 1157–1165. [Google Scholar] [CrossRef]
- Chen, S.; Lin, J.; Liu, H.; Gong, Z.; Wang, X.; Li, M.; Aharoni, A.; Yang, Z.; Yu, X. Insights into Tissue-specific Specialized Metabolism in Tieguanyin Tea Cultivar by Untargeted Metabolomics. Molecules 2018, 23, 1817. [Google Scholar] [CrossRef]
- Wang, W.X.; Zhang, S.W.; Lv, L.S.; Sang, S.M. A new method to prepare and redefine black tea thearubigins. J. Chromatogr. A 2018, 1563, 82–88. [Google Scholar] [CrossRef]
- Li, J.; He, M.; Feng, Y.; Li, T.; Li, Y.; Wu, B.; Ouyang, H.; Yang, S. In-vivo Studies on Metabolism of Luteolin and Luteoloside in Rats. Tradit. Chin. Drug Res. Clin. Plarmacol. 2017, 28, 61–68. [Google Scholar]
- Kalogiouri, N.P.; Aalizadeh, R.; Thomaidis, N.S. Application of an advanced and wide scope non-target screening workflow with LC-ESI-QTOF-MS and chemometrics for the classification of the Greek olive oil varieties. Food Chem. 2018, 256, 53–61. [Google Scholar] [CrossRef]
- Ji, X.Y.; Liu, X.Q.; Peng, Y.X.; Zhan, R.T.; Xu, H.; Ge, X.J. Comparative analysis of methicillin-sensitive and resistant Staphylococcus aureus exposed to emodin based on proteomic profiling. Biochem. Biophys. Res. Commun. 2017, 494, 318–324. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.F.; Wu, S.Y.; Wang, M.; Tian, Y.; Zhang, Z.J.; Song, R. Intestinal metabolism of Polygonum cuspidatum in vitro and in vivo. Biomed. Chromatogr. 2018, 32, e4190. [Google Scholar] [CrossRef] [PubMed]
- Vicente, O.; Boscaiu, M. Flavonoids: Antioxidant Compounds for Plant Defence... and for a Healthy Human Diet. Not. Bot. Horti Agrobot. Cluj-Napoca 2018, 46, 14–21. [Google Scholar] [CrossRef]
- Liu, S.; Song, F.; Liu, Z. Mass Spectrometry Analysis of Traditional Chinese Medicine, 1st ed.; Science China Press: Beijing, China, 2012. [Google Scholar]
- Wu, W.; Liu, Z.Q.; Song, F.R.; Liu, S.Y. Structural analysis of selected characteristic flavones by electrospray tandem mass spectrometry. Anal. Sci. 2004, 20, 1103–1105. [Google Scholar] [CrossRef]
- He, M.; Wu, H.; Nie, J.; Yan, P.; Yang, T.B.; Yang, Z.Y.; Pei, R. Accurate recognition and feature qualify for flavonoid extracts from Liang-wai Gan Cao by liquid chromatography-high resolution-mass spectrometry and computational MS/MS fragmentition. J. Pharm. Biomed. Anal. 2017, 146, 37–47. [Google Scholar] [CrossRef]
- Mekawy, A.M.M.; Abdelaziz, M.N.; Ueda, A. Apigenin pretreatment enhances growth and salinity tolerance of rice seedlings. Plant Physiol. Biochem. 2018, 130, 94–104. [Google Scholar] [CrossRef]
- Karamac, M.; Orak, H.H.; Amarowicz, R.; Orak, A.; Piekoszewski, W. Phenolic contents and antioxidant capacities of wild and cultivated white lupin (Lupinus albus L.) seeds. Food Chem. 2018, 258, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Nabavi, S.F.; Braidy, N.; Gortzi, O.; Sobarzo-Sanchez, E.; Daglia, M.; Skalicka-Wozniak, K.; Nabavi, S.M. Luteolin as an anti-inflammatory and neuroprotective agent: A brief review. Brain Res. Bull. 2015, 119, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kashyap, D.; Sharma, A.; Tuli, H.S.; Sak, K.; Garg, V.K.; Buttar, H.S.; Setzer, W.N.; Sethi, G. Apigenin: A natural bioactive flavone-type molecule with promising therapeutic function. J. Funct. Foods 2018, 48, 457–471. [Google Scholar] [CrossRef]
- Wang, T.Y.; Li, Q.; Bi, K.S. Bioactive flavonoids in medicinal plants: Structure, activity and biological fate. Asian J. Pharm. Sci. 2018, 13, 12–23. [Google Scholar] [CrossRef]
- Ni, J.; Dong, L.X.; Jiang, Z.F.; Yang, X.L.; Sun, Z.H.; Li, J.X.; Wu, Y.H.; Xu, M.J. Salicylic acid-induced flavonoid accumulation in Ginkgo biloba leaves is dependent on red and far-red light. Ind. Crop. Prod. 2018, 118, 102–110. [Google Scholar] [CrossRef]
- Karakaya, S.; Koca, M.; Yılmaz, S.V.; Yıldırım, K.; Pınar, N.M.; Demirci, B.; Brestic, M.; Sytar, O. Molecular Docking Studies of Coumarins Isolated from Extracts and Essential Oils of Zosima absinthifolia Link as Potential Inhibitors for Alzheimer’s Disease. Molecules 2019, 24, 722. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.-S.; Cai, H.; Xue, W.; Wang, M.; Xia, T.; Li, W.-J.; Xing, J.-Q.; Zhao, M.; Huang, Y.-J.; Chen, S.; et al. G3BP1 promotes DNA binding and activation of cGAS. Nat. Immunol. 2018, 20, 18–28. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Huang, Y.; Lian, M.; Fan, Z.; Tian, Y.; Wang, Y.; Kang, H.; Liu, S.; Liu, S.; Li, T.; et al. Metabolic profiling of hepatitis B virus-related hepatocellular carcinoma with diverse differentiation grades. Oncol. Lett. 2017, 13, 1204–1210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nickler, M.; Ottiger, M.; Steuer, C.; Huber, A.; Anderson, J.B.; Mueller, B.; Schuetz, P. Systematic review regarding metabolic profiling for improved pathophysiological understanding of disease and outcome prediction in respiratory infections. Resp. Res. 2015, 16, 125. [Google Scholar] [CrossRef] [Green Version]
- Mo, D.; Mingming, S.; Xianfu, G.; Tie, Z.; Aihua, Z.; Guoxiang, X.; Yunping, Q.; Mingmei, Z.; Zhong, L.; Wei, J. Metabolite profiling of Panax notoginseng using UPLC-ESI-MS. Phytochemistry 2008, 69, 2237–2244. [Google Scholar]
- Li, P.; AnandhiSenthilkumar, H.; Wu, S.-B.; Liu, B.; Guo, Z.-Y.; Fata, J.E.; Kennelly, E.J.; Long, C.-L. Comparative UPLC-QTOF-MS-based metabolomics and bioactivities analyses of Garcinia oblongifolia. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2016, 1011, 179–195. [Google Scholar] [CrossRef]
- Benton, H.P.; Ivanisevic, J.; Mahieu, N.G.; Kurczy, M.E.; Johnson, C.H.; Franco, L.; Rinehart, D.; Valentine, E.; Gowda, H.; Ubhi, B.K.; et al. Autonomous Metabolomics for Rapid Metabolite Identification in Global Profiling. Anal. Chem. 2015, 87, 884–891. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |

| Identification | Precursor Ion | Molecular Formula | Predicted m/z | Measured m/z | PPM | RT | ms2 Fragment Ions at Negative Mode | References |
|---|---|---|---|---|---|---|---|---|
| Tartaric acid | [M − H]− | C4H5O6 | 149.0086 | 149.0082 | 2.68 | 0.72 | 130.9882(2.3), 105.0215(5.60),103.0071(1.41), 87.0133(36.37), 72.9965(100) | HMDB, METLIN |
| Malic acid | [M − H]− | C4H5O5 | 133.0137 | 133.0133 | 3.01 | 0.72 | 115.0076(19.99), 89.0280(7.69), 72.9970(37.65), 71.0176(100),59.0163(7.95) | HMDB, METLIN |
| Citric acid | [M − H]− | C6H7O7 | 191.0192 | 191.0201 | 4.71 | 0.80 | 173.0125(3.75), 111.0125(100), 87.0126(65.85), 85.0336(76.01), 67.0217(21.87), 57.0419(17.03)103.0106 | HMDB, METLIN |
| Gallic acid | [M − H]− | C7H5O5 | 169.0215 | 169.0218 | 1.77 | 0.97 | 125.0284(100), 107.0178 (8.69), 111.0112(3.30), 97.0326(14.68), 79.0236(23.64), 69.0375(19.23) | Standard, METLIN |
| 6-Galloylglucose/2-Galloylglucose | [M − H]− | C13H15O10 | 331.0663 | 331.0664 | 0.30 | 1.14 | 241.0128(3.41), 211.0290(13.62), 169.0172(100), 151.0054(48.81), 137.0258(4.43),125.0263(43.17), 111.0117(17.48) | HMDB, [2] |
| Salicylic acid beta-d-glucoside | [M − H]− | C13H15O8 | 299.0772 | 299.0775 | 1.00 | 1.34 | 225.0779(7.62), 179.0422(46.39), 137.0294(100), 119.0522(8.48), 93.0370(10.73), 73.0299(23.70),71.0166(26.89) | HMDB, METLIN |
| Epiafzelechin 3-gallate | [M − H]− | C22H17O9 | 425.0873 | 425.0861 | 2.82 | 2.23 | 407.0737(100), 255.0812(4.98), 151.0423(36.55), 137.0292(64.75), 125.0224(11.75) | HMDB |
| Epigallocatechin/Gallocatechin | [M − H]− | C15H13O7 | 305.0661 | 305.0672 | 3.61 | 2.31 | 219.0648(56.12), 191.0689(34.03), 167.0307(52.37), 139.0480(29.64), 137.0315(30.43), 125.0235(100), 111.0429(30.40) | HMDB, METLIN |
| Glucocaffeic acid | [M − H]− | C15H17O9 | 341.0873 | 341.0886 | 3.81 | 2.59 | 221.0438, 179.0379(100), 161.0244(6.52), 151.0395(11.84), 135.0485(33.41) | HMDB |
| Catechin | [M − H]− | C15H13O6 | 289.0712 | 289.0712 | 0.00 | 2.64 | 271.0601(5.61), 245.0827(23.84), 203.0720(53.62), 151.0424(28.46), 109.0320(100) | HMDB, Standard, [15,21] |
| Chlorogenic acid | [M − H]− | C16H17O9 | 353.0852 | 353.0845 | 1.98 | 2.83 | 191.0585(100), 179.0378(5.08), 161.0270(2.69), 87.0068(2.1) | HMDB, METLIN, [19] |
| Epigallate catechin gallate | [M − H]− | C22H17O11 | 457.0771 | 457.0759 | 2.63 | 2.93 | 303.0573(100), 287.0546(15.4), 169.0170(8.76), 165.0557(12.6) 161.0138(3.66), 151.0047(75.84), 125.0271(32.68) | HMDB, METLIN, |
| 3-p-Coumaroylquinic acid | [M − H]− | C16H17O8 | 337.0923 | 337.0931 | 2.37 | 3.29 | 277.0685(10.43), 231.0627(2.99), 191.0540(29.44), 173.0443(100), 163.0440(26.17), 119.0509(13.82) | HMDB |
| Epicatechin | [M − H]− | C15H13O6 | 289.0712 | 289.0707 | 1.73 | 3.59 | 289.0729(53.01), 245.0939(33.89), 221.0838(18.71), 203.0728(100), 151.0438(45.34), 123.0561(73.41), 109.0356(66.28) | HMDB, [2,21] |
| Apigenin-6-glucoside 8-carabinoside | [M − H]− | C26H27O14 | 563.1395 | 563.1385 | 1.77 | 4.12 | 563.1375(100), 545.1283(3.52), 503.1142(7.92), 473.1088(10.53), 443.0946(12.11), 383.0897(10.82), 353.0642(10.79) | HMDB, [22] |
| Procyanidin B1 | [M − H]− | C30H25O12 | 577.1346 | 577.1371 | 4.33 | 4.36 | 559.1370, 535.1225(23.27), 425.0878(38.95), 289.0731(100), 125.0276(54.62) | HMDB, [2,21] |
| 3-galloylprocyanidin B1/B2 | [M − H]− | C37H29O16 | 729.1456 | 729.1429 | 3.70 | 4.42 | 603.1210(10.82), 577.1231(32.4), 441.0865(19.36), 407.0865(100), 289.0757(40.38), 271.0661(8.43), 125.0244(18.96) | HMDB, METLIN |
| Resveratrol-4′-o-d-glucopyranoside | [M + COOH]- | C21H23O10 | 435.1286 | 435.1290 | 0.92 | 4.67 | 389.1263(2.05), 227.0733(100), 185.0636(1.17), 143.0524(0.64), | [2,21] |
| Epicatechin gallate | [M − H]− | C22H17O11 | 441.0822 | 441.0809 | 2.95 | 4.69 | 303.0592(2.11),290.0796(14.52), 289.0770(47.78), 271.0629(7.1), 245.0826(8.03),169.0182(100), 151.0501(2.87), 125.0281(26.39) | HMDB, [21,22,23] |
| Piceid | [M − H]− [M + HCOO]- | C20H21O8 C21H23O10 | 389.1240 435.1286 | 389.1236 435.1279 | 1.03 1.61 | 5.09 5.09 | 227.0723(100), 185.0687(5.44), 143.0526(1.28), 389.1263(2.05), 227.0733(100) | HMDB, METLIN, Standard, [21] |
| Rutin | [M − H]− | C27H29O16 | 609.1456 | 609.1480 | 3.94 | 5.09 | 609.1493(100), 343.0538(1.34), 301.0387(16.57), 300.0319(25.44), 271.0245(1.43), 255.0305(0.78), 151.0107(0.63), | HMDB, METLIN, [23] |
| Catechin gallate | [M − H]− | C22H17O10 | 441.0822 | 441.0809 | 2.95 | 5.23 | 303.0549(1.63), 289.0750(53.51), 245.0450(13.48), 205.0571(3.22), 169.0179(100), | HMDB, [22] |
| Quercetin 4′-glucoside | [M − H]− | C21H19O12 | 463.0877 | 463.0859 | 3.87 | 5.30 | 343.0383(1.45), 301.0367(100), 283.0280 (1.21), 178.9999(6.03), 151.0080(2.21), 107.0185(0.65) | HMDB, METLIN |
| kaempferol galactoside | [M − H]− | C21H19O11 | 447.0922 | 447.0940 | 4.03 | 5.36 | 285.0455(100), 284.0392(17.21), 245.088(32.35) | HMDB, [22] |
| Resveratrol-3-o-beta-d-(sulfonyl)-glucopyranoside | [M − H]− | C20H21O11S | 469.0799 | 469.0776 | 4.70 | 5.48 | 269.0746(4.56), 227.0669(8.43), 241.0073(100) | HMDB, [2,21] |
| Quercetin xyloside | [M − H]− | C20H17O11 | 433.0771 | 433.0759 | 2.77 | 5.88 | 301.0384(100), 283.0242(1.57), 271.0315(11.66), 243.0355(1.93), 151.0068(5.46) | HMDB |
| Hydroxyl aloe-emodin-o-glucoside | [M − H]− | C21H19O9 | 447.0932 | 447.0949 | 3.80 | 6.03 | 241.0542(100), 403.1076(31.52), 197.0674(6.79), 161.0422(1.32) | [2], HMDB, METLIN, [21] |
| Resveratrol-3-d-(6 or 2-galloyl)glucopyranoside | [M − H]− | C27H25O12 | 541.1341 | 541.1337 | 0.74 | 6.17 | 541.1337(100), 417.093(9.36), 313.0581(25.1), 227.0760(10.23), 169.0157(13.53) | [2], METLIN |
| Procyanidin-B-1, 3-o-gallate | [M − H]− | C37H29O16 | 729.1450 | 729.1463 | 1.78 | 6.24 | 245.0816(19.85), 289.0719(59.57), 407.0833(100), 451.0952(26.42) | [2], HMDB |
| Kaempferide | [M − H]− | C16H11O6 | 299.0556 | 299.0566 | 3.34 | 6.43 | 283.0082(4.81), 271.0251(6.79), 213.0597(100), 151.0082(17.20) | HMDB |
| Apigenin 7-O-glucoside | [M − H]− | C21H19O10 | 431.0978 | 431.0995 | 3.94 | 6.69 | 431.1004(100), 311.0561(0.65), 269.0497(92.44), 240.0466(10.54) | HMDB [2], METLIN |
| Resveratrol | [M − H]− | C14H11O3 | 227.0708 | 227.0715 | 3.08 | 6.95 | 211.0384(6.59), 185.0640(100), 183.0846(10.63) 159.0824(11.21), 143.0506(80.26), 133.0355 | Standard, HMDB, METLIN, [18] |
| Emodin-o-(sulfonyl)-glucoside | [M − H]− | C21H19O13S | 511.0541 | 511.0547 | 1.17 | 7.21 | 431.1007(100), 311.0631(1.68), 270.0539(12.13), 269.0485(41.04), 257.0494(1.03), 241.0040(4.82), 225.0545(0.82) | [2], METLIN, [21] |
| Torachrysone 8-glucoside | [M − H]− | C20H23O9 | 407.1342 | 407.1353 | 2.70 | 7.86 | 245.0811(100), 269.0744(0.4) | METLIN, HMD, [21] |
| Luteolin | [M − H]− | C15H9O6 | 285.0394 | 285.0387 | 2.46 | 8.04 | 285.0381(100), 267.0342(2.78), 257.0439(1.03),243.0279(1.84), 241.0489(2.14), 229.0493, 217.0488(3.9), 175.0381(5.29), 151.0031(11.51), 133.0329(18.27), 121.0273(0.73), 107.0162(3.31) | HMDB, METLIN, [2,24] |
| Emodin-6-o-glucoside | [M − H]− | C21H19O10 | 431.0978 | 431.0982 | 0.93 | 8.14 | 431.1000(48.67), 311.0584(4.54), 293.0518(1.66), 282.0597(0.7), 269.0497(100), 225.0582(2.41), | HMDB, [21] |
| Torachrysone-8-o-(sulfonyl)-glucoside | [M − H]− | C20H23O12S | 487.0905 | 487.0892 | 2.67 | 8.41 | 407.1355, 287.0927(1.03), 245.0841(100), 243.0051(8.32), 241.0076(90.07), 230.0601(7.32), 215.0435(0.55), 113.0251(1.32) | [2,21,22] |
| Hydroxyl aloe-emodin | [M − H]− | C15H9O6 | 285.0394 | 285.0387 | 2.46 | 8.93 | 285.0401(100), 241.0543(2.17), 211.0421(0.34), 197.0586(0.69), 195.0533(0.4), 167.05(0.33), 151.0073(10.22), 133.0319(14.97), | HMDB, [21] |
| 6″-Malonylgenistin | [M − H]− | C24H21O13 | 517.0982 | 517.0983 | 019 | 9.05 | 473.1056(100), 455.0952(0.53), 431.0968(2.51), 311.0553(3.47), 269.0454(75.44), 413.0835(0.25) | [22], HMDB |
| Apigenin | [M − H]− | C15H9O5 | 269.0450 | 269.0449 | 0.37 | 9.10 | 269.0494(100), 241.0493(2.97), 227.0409(3.04), 225.0536(36.73), 151.0063(1.39), | METLIN, HMDB, [25] |
| Apigenin 7-O-(6″-O-acetylglucoside) | [M − H]− | C23H21O11 | 473.1084 | 473.1086 | 0.42 | 9.15 | 473.1107(100), 431.0898(10.92), 413.0669(10.97), 311.0626(16.45), 269.0460(37.55), | HMDB |
| Bergapten | [M − H]− | C12H7O4 | 215.0344 | 215.0351 | 3.26 | 9.23 | 171.0469(2.83), 159.0484(100), 143.0535(18.1), 131.0531(86.87) | HMDB, METLIN |
| Torachrysone-8-o-(acetyl)-glucoside | [M − H]− | C22H25O10 | 449.1442 | 449.1439 | 0.45 | 9.24 | 269.0438(2.47), 245.0821(100), 230.0583(11.11), 215.0363(2.21), | [2], HMDB |
| Acetylemodin-o-glucoside | [M − H]− | C23H21O11 | 473.1078 | 473.1073 | 1.06 | 9.54 | 335.9747(3.94), 311.0626(16.45), 283.0506(5.9), 269.0460(100), 267.07(12.37) | [2,21] |
| Physcion-8-glucoside | [M − H]− | C22H21O10 | 445.1129 | 445.1145 | 3.59 | 9.71 | 325.0434(2.31), 283.0630(100), 240.0445(14.86), 165.0464(1.63), | [18,21] |
| 6″-O-Acetylglycitin | [M − H]− | C24H23O11 | 487.1240 | 487.1240 | 0 | 11.35 | 283.0645(100), 267.0868(1.18) | HMDB |
| 6-hydroxylrhein | [M − H]− | C15H7O7 | 299.0186 | 299.0187 | 0.33 | 11.48 | 300.0192(32.33), 299.0193(100), 255.0328(16.88), 227.0381(6.13), 211.0423(12.13), 199.0473(1.99) | [2,21] |
| Formononetin 7-O-glucoside | [M − H]− | C22H21O9 | 429.1186 | 429.1175 | 2.56 | 12.18 | 411.0650(7.35), 307.0586(7.87), 293.0452(100), 267.0667(21.06), 237.0614(6.34) | HMDB, METLIN |
| Acetylemodin | [M − H]− | C17H11O6 | 311.0551 | 311.0560 | 3.11 | 12.79 | 311.0563(100), 269.0429(14.3), 268.0402(48.88), 240.0466(3.84), 224.0481(1.22),196.0565(1.02) | [2,21] |
| Apigenin 7-sulfate | [M − H]− | C15H9O8S | 349.0018 | 349.0034 | 4.58 | 12.81 | 331.2035(0.19), 306.3984(0.17), 269.0437(100), 241.0124(0.71), 227.0115(0.45) | HMDB, METLIN |
| Emodin | [M − H]− | C15H9O5 | 269.0450 | 269.0460 | 3.71 | 15.00 | 241.0512(21.33), 225.0546(100), 197.0626(1.11), 181.0666(1.33), 157.0626(0.39), 65.0044(0.14) | Standard, [18], HMDB, [21] |
| Physcion | [M − H]− | C16H11O5 | 283.0606 | 283.0611 | 1.77 | 15.39 | 269.0381(24.64), 268.0362(100), 241.0456(8.91), 240.0412(21.34), 205.0512(13.4), 197.2618(1.65) | METLIN, [21] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Z.; Wang, X.; Chen, M.; Hu, H.; Cao, J.; Chai, T.; Wang, H. A Study on Tissue-Specific Metabolite Variations in Polygonum cuspidatum by High-Resolution Mass Spectrometry-Based Metabolic Profiling. Molecules 2019, 24, 1058. https://doi.org/10.3390/molecules24061058
Wu Z, Wang X, Chen M, Hu H, Cao J, Chai T, Wang H. A Study on Tissue-Specific Metabolite Variations in Polygonum cuspidatum by High-Resolution Mass Spectrometry-Based Metabolic Profiling. Molecules. 2019; 24(6):1058. https://doi.org/10.3390/molecules24061058
Chicago/Turabian StyleWu, Zhijun, Xiaowei Wang, Mo Chen, Hongyan Hu, Jie Cao, Tuanyao Chai, and Hong Wang. 2019. "A Study on Tissue-Specific Metabolite Variations in Polygonum cuspidatum by High-Resolution Mass Spectrometry-Based Metabolic Profiling" Molecules 24, no. 6: 1058. https://doi.org/10.3390/molecules24061058






