Practical Microwave Synthesis of Carbazole Aldehydes for the Development of DNA-Binding Ligands
Abstract
:1. Introduction
2. Results and Discussion
2.1. Chemistry
2.2. Spectral Properties of Ligand 3c
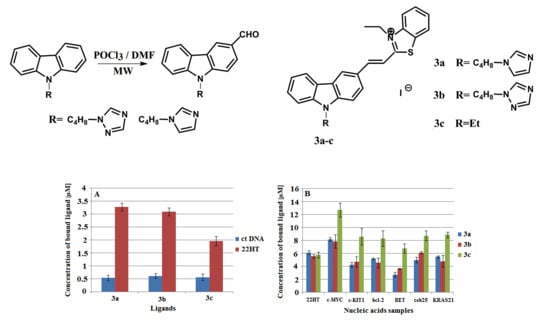
2.3. DNA Binding
3. Materials and Methods
3.1. General Information
3.2. Materials
3.2.1. General Procedure of Synthesis of Compounds 2a and 2b
3.2.2. Synthesis of 3-[(3-ethyl)-2-vinylbenzothiazolium]-9-N-ethyl carbazole iodide 3c
3.3. Methods
3.3.1. Absorption and Fluorescence Spectroscopy
3.3.2. Dialysis Assay Method
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Parkinson, G.N.; Lee, M.P.; Neidle, S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature 2002, 417, 876–880. [Google Scholar] [CrossRef] [PubMed]
- Mathad, R.I.; Hatzakis, E.; Dai, J.; Yang, D. c-MYC promoter G-quadruplex formed at the 5′-end of NHE III1 element: Insights into biological relevance and parallel-stranded G-quadruplex stability. Nucleic Acids Res. 2011, 39, 9023–9033. [Google Scholar] [CrossRef] [PubMed]
- Rankin, S.; Reszka, A.P.; Huppert, J.; Zloh, M.; Parkinson, G.N.; Todd, A.K.; Ladame, S.; Balasubramanian, S.; Neidle, S. Putative DNA quadruplex formation within the human c-kit oncogene. J. Am. Chem. Soc. 2005, 127, 10584–10589. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, P.; Lin, C.; Mathad, R.I.; Carver, M.; Yang, D. The Major G-Quadruplex Formed in the Human BCL-2 Proximal Promoter Adopts a Parallel Structure with a 13-nt Loop in K+ Solution. J. Am. Chem. Soc. 2014, 136, 1750–1753. [Google Scholar] [CrossRef] [PubMed]
- Tong, X.; Lan, W.; Zhang, X.; Wu, H.; Liu, M.; Cao, C. Solution structure of all parallel G-quadruplex formed by the oncogene RET promoter sequence. Nucleic Acids Res. 2011, 39, 6753–6763. [Google Scholar] [CrossRef] [PubMed]
- Biffi, G.; Tannahill, D.; McCafferty, J.; Balasubramanian, S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013, 5, 182–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ou, T.-M.; Lu, Y.-J.; Tan, J.-H.; Huang, Z.-S.; Wong, K.-Y.; Gu, L.-G. G-Quadruplexes: Targets in Anticancer Drug Design. ChemMedChem 2008, 3, 690–713. [Google Scholar] [CrossRef]
- De Cian, A.; Lacroix, L.; Douarre, C.; Temime-Smaali, N.; Trentesaux, C.; Riou, J.-F.; Mergny, J.-L. Targeting telomeres and telomerase. Biochemie 2008, 90, 131–155. [Google Scholar] [CrossRef]
- Ramos, C.I.V.; Almeida, S.P.; Lourenço, L.M.O.; Pereira, P.M.R.; Fernandes, R.; Faustino, M.A.F.; Tomé, J.P.C.; Carvalho, J.; Cruz, C.; Neves, M.G.P.M.S. Multicharged Phthalocyanines as Selective Ligands for G-Quadruplex DNA structures. Molecules 2019, 24, 733. [Google Scholar] [CrossRef]
- Ohnmacht, S.A.; Neidle, S. Small-molecule quadruplex-targeted drug discovery. Bioorg. Med. Chem. Lett. 2014, 24, 2602–2612. [Google Scholar] [CrossRef]
- Neidle, S. Quadruplex nucleic acids as targets for anticancer therapeutics. Nat. Rev. Chem. 2017, 1, 0041. [Google Scholar] [CrossRef]
- Asamitsu, S.; Bando, T.; Sugiyama, H. Ligand Design to Acquire Specificity to Intended G-Quadruplex Structures. Chem. Eur. J. 2019, 25, 417–430. [Google Scholar] [CrossRef] [PubMed]
- Głuszyńska, A.; Rajczak, E.; Juskowiak, B. Synthesis and spectroscopic characterisation of (E)-2-(2-(9-(4-(1H-1,2,4-triazol-1-yl)butyl)-9H-carbazol-3-yl)vinyl)-3-ethylbenzo-[d]thiazol-3-ium, a new ligand and potential DNA intercalator. Chem. Pap. 2013, 67, 1231–1239. [Google Scholar] [CrossRef]
- Głuszyńska, A.; Juskowiak, B.; Kuta-Siejkowska, M.; Hoffmann, M.; Haider, S. Carbazole ligands as c-myc G-quadruplex binders. Int. J. Biol. Macromol. 2018, 114, 479–490. [Google Scholar] [CrossRef]
- Głuszyńska, A.; Juskowiak, B.; Kuta-Siejkowska, M.; Hoffmann, M.; Haider, S. Carbazole derivatives’ binding to c-KIT G-quadruplex DNA. Molecules 2018, 23, 1134. [Google Scholar] [CrossRef] [PubMed]
- Głuszyńska, A.; Juskowiak, B.; Rubiś, B. Binding study of the fluorescent carbazole derivative with human telomeric G-quadruplexes. Molecules 2018, 23, 3154. [Google Scholar] [CrossRef] [PubMed]
- Głuszyńska, A. Biological potential of carbazole derivatives. Eur. J. Med. Chem. 2015, 94, 405–426. [Google Scholar] [CrossRef]
- Zhang, L.; Peng, X.-M.; Damu, G.L.V.; Geng, R.-X.; Zhou, C.-H. Comprehensive review in current developments of imidazole-based medicinal chemistry. Med. Res. Rev. 2014, 34, 340–437. [Google Scholar] [CrossRef]
- Kaur, R.; Dwivedi, A.R.; Kumar, B.; Kumar, V. Recent Developments on 1,2,4-Triazole Nucleus in Anticancer Compounds: A Review. Anticancer Agents Med. Chem. 2016, 16, 465–489. [Google Scholar] [CrossRef]
- Kim, H.K.; Ryu, M.-K.; Kim, K.-D.; Lee, S.-M.; Cho, S.-W.; Park, J.-W. Tunable electroluminescence from silicon-containing poly(p-phenylenevinylene)-related copolymers with well-defined structures. Macromolecules 1998, 31, 1114–1123. [Google Scholar] [CrossRef]
- Song, Y.; Di, C.-A.; Wei, Z.; Zhao, T.; Xu, W.; Liu, Y.; Zhang, D.; Zhu, D. Synthesis, characterization, and fieldeffect transistor properties of carbazolenevinylene oligomers: From linear to cyclic architectures. Chem. Eur. J. 2008, 14, 4731–4740. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Chen, G.; Xu, X.; Chen, H.; Ji, S. The synthesis and optical properties of benzothiazole-based derivatives with various π-electron donors as novel bipolar fluorescent compounds. Dyes Pigments 2010, 86, 238–248. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, L.; Wada, T.; Sasabe, H. Synthesis and characterization of novel hyperbranched polymer with dipole carbazole moieties for multifunctional materials. J. Polym. Sci. A Polym. Chem. 1996, 34, 1359–1363. [Google Scholar] [CrossRef] [Green Version]
- Xu, T.H.; Lu, R.; Qiu, X.P.; Liu, X.L.; Xue, P.C.; Tan, C.H.; Bao, C.Y.; Zhao, Y.Y. Synthesis and characterization of carbazole-based dendrimers with porphyrin cores. Eur. J. Org. Chem. 2006, 2006, 4014–4020. [Google Scholar] [CrossRef]
- Ryu, H.; Subramanian, L.R.; Hanack, M. Photo and electroluminescent properties of cyano-substituted styryl derivatives and synthesis of CN–PPV model compounds containing an alkoxy spacer for OLEDs. Tetrahedron 2006, 62, 6236–6247. [Google Scholar] [CrossRef]
- Fei, X.; Gu, Y.; Li, C.; Yang, X. Study on Synthesis and Spectrum of Novel Styryl Cyanine Dyes with a Carbazole Bridged Chain. J. Fluoresc. 2012, 22, 807–814. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.-F.; Gan, L.-L.; Zhou, C.-H. Synthesis, antibacterial and antifungal activities of some carbazole derivatives. Bioorg. Med. Chem. Lett. 2010, 20, 1881–1884. [Google Scholar] [CrossRef]
- Loupy, A. Microwave in Organic Synthesis, 2nd ed.; Wiley-WCH: Weinheim, Germany, 2006. [Google Scholar]
- Elgemeie, G.H.; Mohamed, R.A. Microwave synthesis of fluorescent and luminescent dyes (1990–2017). J. Mol. Struct. 2018, 1173, 707–742. [Google Scholar] [CrossRef]
- Hall, H.K., Jr. Correlation of the Base Strengths of Amines. J. Am. Chem. Soc. 1957, 79, 5441–5444. [Google Scholar] [CrossRef]
- Saengkhae, C.; Salerno, M.; Adès, D.; Siove, A.; Le Moyec, L.; Migonney, V.; Garnier-Suillerot, A. Ability of carbazole salts, inhibitors of Alzheimer β-amyloid fibril formation, to cross cellular membranes. Eur. J. Pharmacol. 2007, 559, 124–131. [Google Scholar] [CrossRef]
- Lin, D.; Fei, X.; Gu, Y.; Wang, C.; Tang, Y.; Li, R.; Zhou, J. A benzindole substituted carbazole cyanine dye: A novel targeting fluorescent probe for parallel c-myc G-quadruplexes. Analyst 2015, 140, 5772–5780. [Google Scholar] [CrossRef] [PubMed]
- Murat, P.; Singh, Y.; Defrancq, E. Methods for investigating G-quadruplex DNA/ligand interactions. Chem. Soc Rev. 2011, 40, 5293–5307. [Google Scholar] [CrossRef] [PubMed]
- Jaumot, J.; Gargallo, R. Experimental Methods for Studying the Interactions between G-Quadruplex Structures and Ligands. Curr. Pharm. Des. 2012, 18, 1900–1916. [Google Scholar] [CrossRef] [PubMed]
- Musumeci, D.; Amato, J.; Randazzo, A.; Novellino, E.; Giancola, C.; Montesarchio, D.; Pagano, B. G-Quadruplex on Oligo Affinity Support (G4-OAS): An Easy Affinity Chromatography-Based Assay for the Screening of G-Quadruplex Ligands. Anal. Chem. 2014, 86, 4126–4130. [Google Scholar] [CrossRef] [PubMed]
- Ragazzon, P.A.; Chaires, J.B. Use of competition dialysis in the discovery of G-quadruplex selective ligands. Methods 2007, 43, 313–323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ragazzon, P.A.; Garbett, N.C.; Chaires, J.B. Competition dialysis: A method for the study of structural selective nucleic acid binding. Methods 2007, 42, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Chaires, J.B. Sequence and Structural Selectivity of Nucleic Acid Binding Ligands. Biochemistry 1999, 38, 16067–16075. [Google Scholar] [CrossRef] [PubMed]
- Guittat, L.; Alberti, P.; Rosu, F.; Van Miert, S.; Thetiot, E.; Pieters, L.; Gabelica, V.; De Pauw, E.; Ottaviani, A.; Riou, J.-F.; et al. Interactions of cryptolepine and neocryptolepine with unusual DNA structures. Biochimie. 2003, 85, 535–547. [Google Scholar] [CrossRef] [Green Version]
- Rosu, F.; De Pauw, E.; Guittat, L.; Alberti, P.; Lacroix, L.; Mailliet, P.; Riou, J.-F.; Mergny, J.-L. Selective interaction of ethidium derivatives with quadruplexes: An equilibrium dialysis and electrospray ionization mass spectrometry analysis. Biochemistry 2003, 42, 10361–10371. [Google Scholar] [CrossRef]
- Granotier, C.; Pennarun, G.; Riou, L.; Hoffschir, F.; Gauthier, L.R.; De Cian, A.; Gomez, D.; Mandine, E.; Riou, J.-F.; Mergny, J.-L.; et al. Preferential binding of a G-quadruplex ligand to human chromosome ends. Nucleic Acids Res. 2005, 33, 4182–4190. [Google Scholar] [CrossRef] [Green Version]
- Ou, T.-M.; Lu, Y.-J.; Zhang, C.; Huang, Z.-S.; Wang, X.-D.; Tan, J.-H.; Chen, Y.; Ma, D.-L.; Wong, K.-Y.; Tang, J.C.-O.; et al. Stabilization of G-Quadruplex DNA and Down-Regulation of Oncogene c-myc by Quindoline Derivatives. J. Med. Chem. 2007, 50, 1465–1474. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Leung, C.-H.; Ma, D.-L.; Yan, S.-C.; Che, C.-M. Structure-Based Design of Platinum(II) Complexes as c-myc Oncogene Down-Regulators and Luminescent Probes for G-Quadruplex DNA. Chem. Eur. J. 2010, 16, 6900–6911. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.; Ranjan, N.; Arya, D.P. Synthesis and spectroscopic studies of the aminoglycoside (Neomycin)-perylene conjugate binding to human telomeric DNA. Biochemistry 2011, 50, 2838–2849. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lu, K.; Xuan, S.; Toh, Z.; Zhang, D.; Shao, F. A Pt(II)–Dip complex stabilizes parallel c-myc G-quadruplex. Chem. Commun. 2013, 42, 4758–4760. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, R.; Aviñó, A.; Pérez-Tomás, R.; Gargallo, R.; Eritja, R.J. Synthesis and G-Quadruplex-Binding Properties of Defined Acridine Oligomers. Nucleic Acids 2010, 2010, 489060. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, R.; Artali, R.; Farrera-Sinfreu, J.; Albericio, F.; Royo, M.; Eritja, R.; Mazzini, S. Acridine and quindoline oligomers linked through a 4-aminoproline backbone prefer G-quadruplex structures. Biochim Biophys Acta 2011, 1810, 769–776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferreira, R.; Artali, R.; Benoit, A.; Gargallo, R.; Eritja, R.; Ferguson, D.M.; Sham, Y.Y.; Mazzini, S. Structure and Stability of Human Telomeric G-Quadruplex with Preclinical 9-Amino Acridines. PLoS ONE 2013, 8, e57701. [Google Scholar] [CrossRef]
- Tataurov, A.V.; You, Y.; Owczarzy, R. Predicting ultraviolet spectrum of single stranded and double stranded deoxyribonucleic acids. Biophys. Chem. 2008, 133, 66–70. [Google Scholar] [CrossRef]
Sample Availability: Not available. |




| Entry | Substrate | Vilsmayer Reagent [equiv.] | T [°C] | t [min] a | Y [%] b |
|---|---|---|---|---|---|
| 1 | 1a | 1 | 100 | 120 | 49 |
| 2 | 1a | 2 | 100 | 120 | 58 |
| 3 | 1a | 3 | 100 | 80 | 82 |
| 4 | 1a | 3 | 100 | 100 | 88 |
| 5 | 1a | 3 | 120 | 60 | 78 |
| 6 | 1a | 4 | 100 | 90 | 58 |
| 7 | 1a | 19 | 100 | 15 | 51 |
| 8 | 1b | 3 | 100 | 100 | 86 |
| Solvent | ε/104 [M−1∙cm−1] | Abs. λmax [nm] | Em. λmax [nm] | Stockes Shift [nm] | Relative Fluorescence Yield [%] |
|---|---|---|---|---|---|
| CH2Cl2 | 5.6 ± 0.1 | 504 | 571 | 67 | 45.5 |
| CHCl3 | 5.5 ± 0.1 | 509 | 567 | 58 | 49.9 |
| MeOH | 5.2 ± 0.1 | 475 | 568 | 93 | 22.2 |
| EtOH | 5.1 ± 0.1 | 480 | 570 | 90 | 26.2 |
| ACN | 4.8 ± 0.1 | 470 | 569 | 99 | 22.3 |
| H2O | 4.3 ± 0.1 | 452 | 568 | 116 | 3.2 |
| DMSO | 4.2 ± 0.1 | 472 | 572 | 100 | 31.2 |
| 10 mM Tris-HCl | 3.9 ± 0.1 | 451 | 567 | 116 | 2.3 |
| Toluene | 3.3 ± 0.1 | 468 | 567 | 99 | 3.7 |
| 1,4-Dioxane | 2.4 ± 0.1 | 483 | 565 | 82 | 42.9 |
| Structure | Sample | DNA or Oligonucleotide | Monomer Unit |
|---|---|---|---|
| Double-stranded | ctDNA | Calf thymus DNA | Base pair |
| Tetra-stranded | 22HT | 5′-AGGGTTAGGGTTAGGGTTAGGG-3′ | quadruplex |
| c-MYC | 5′-TGAGGGTGGGTAGGGTGGGTAA-3′ | ||
| c-KIT1 | 5′-AGGGAGGGCGCTGGGAGGAGGG-3′ | ||
| Bcl-2 | 5′-GGGCGCGGGAGGAATTGGGCGGG-3′ | ||
| RET | 5′-GGGGCGGGGCGGGGCGGGGT-3′ | ||
| ceb25 | 5′-AAGGGTGGGTGTAAGTGTGGGTGGGT-3′ | ||
| KRAS21 | 5′-AGGGCGGTGTGGGAAGAGGGA-3′ |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Głuszyńska, A.; Juskowiak, B. Practical Microwave Synthesis of Carbazole Aldehydes for the Development of DNA-Binding Ligands. Molecules 2019, 24, 965. https://doi.org/10.3390/molecules24050965
Głuszyńska A, Juskowiak B. Practical Microwave Synthesis of Carbazole Aldehydes for the Development of DNA-Binding Ligands. Molecules. 2019; 24(5):965. https://doi.org/10.3390/molecules24050965
Chicago/Turabian StyleGłuszyńska, Agata, and Bernard Juskowiak. 2019. "Practical Microwave Synthesis of Carbazole Aldehydes for the Development of DNA-Binding Ligands" Molecules 24, no. 5: 965. https://doi.org/10.3390/molecules24050965