Bilobalide Suppresses Adipogenesis in 3T3-L1 Adipocytes via the AMPK Signaling Pathway
Abstract
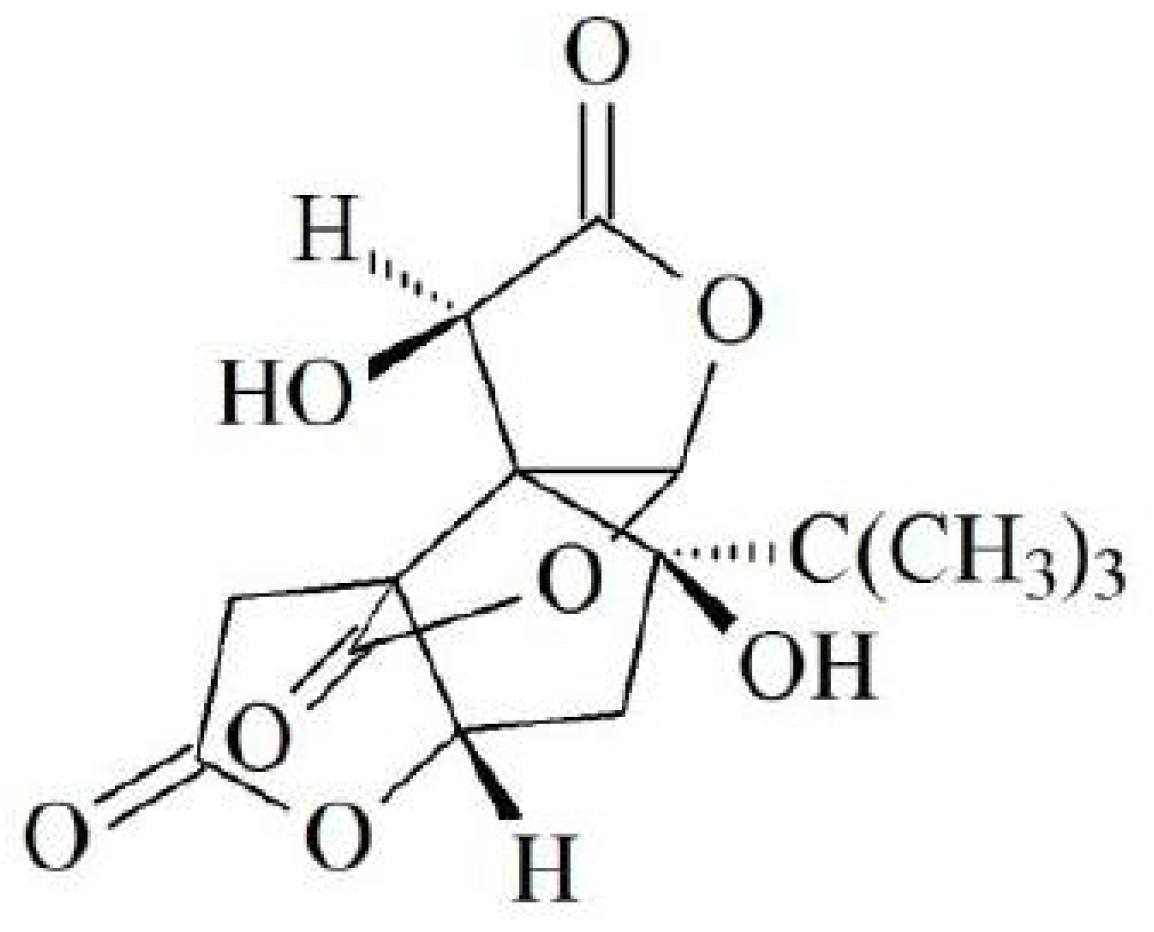
1. Introduction
2. Results
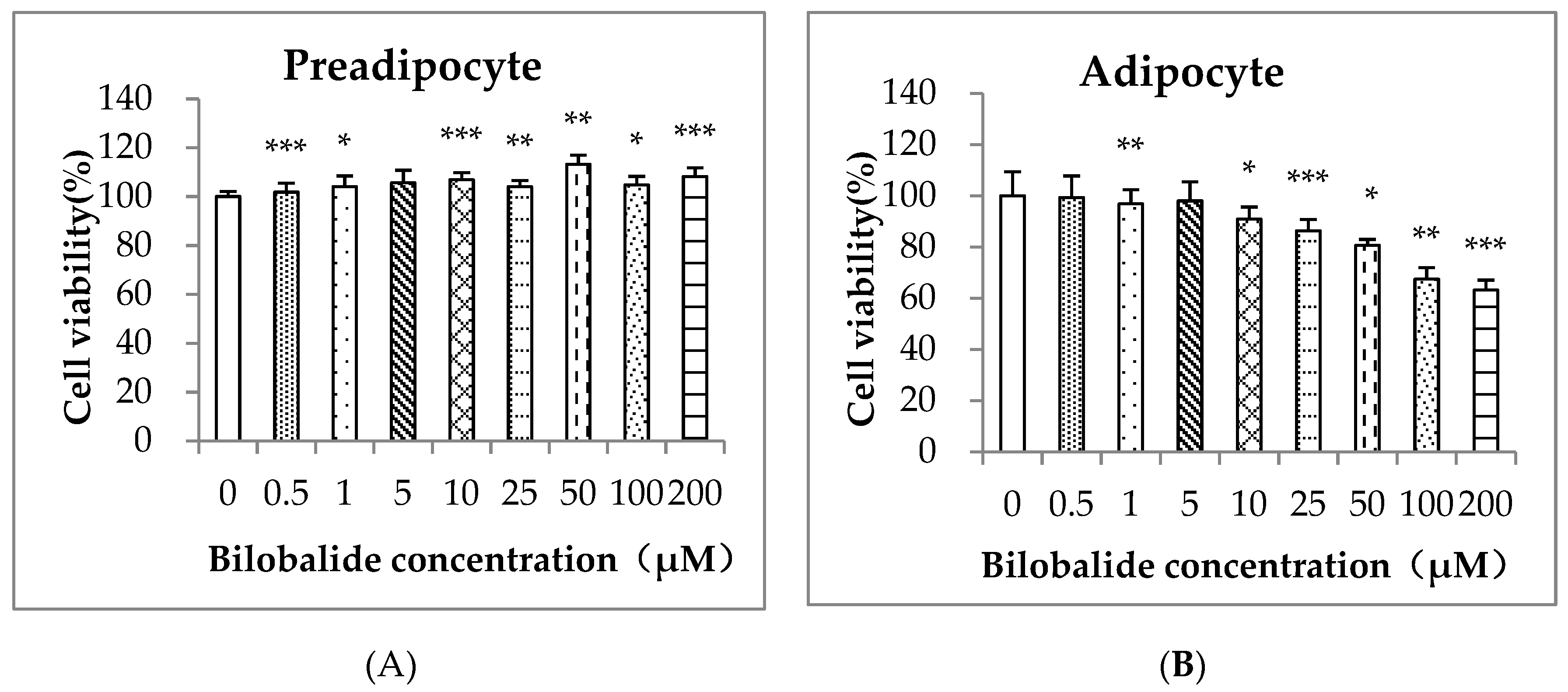
2.1. Cell Cytotoxicity of Bilobalide on 3T3-L1 Cells
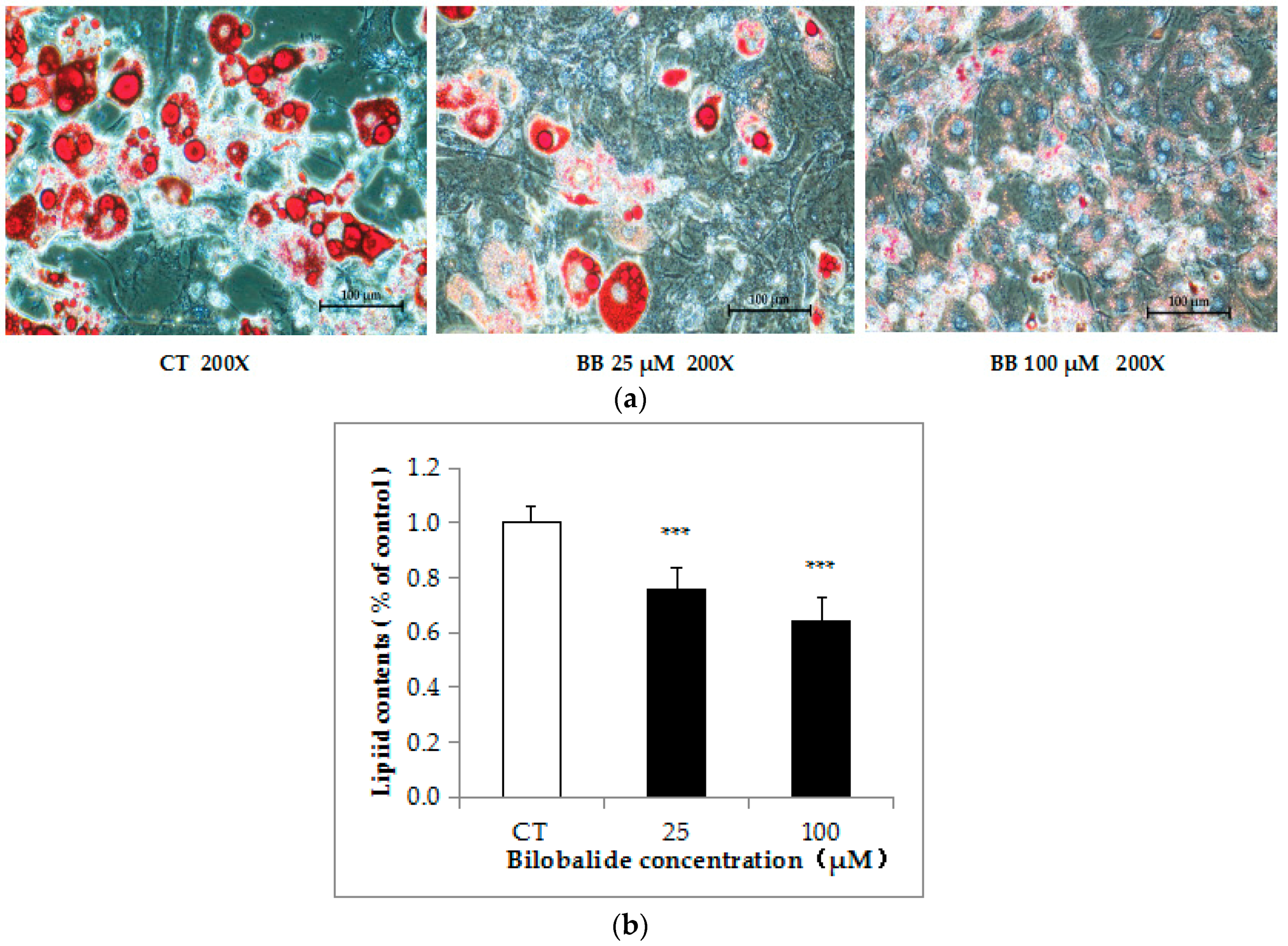
2.2. Bilobalide Inhibits Lipid Accumulation in 3T3-L1 cells
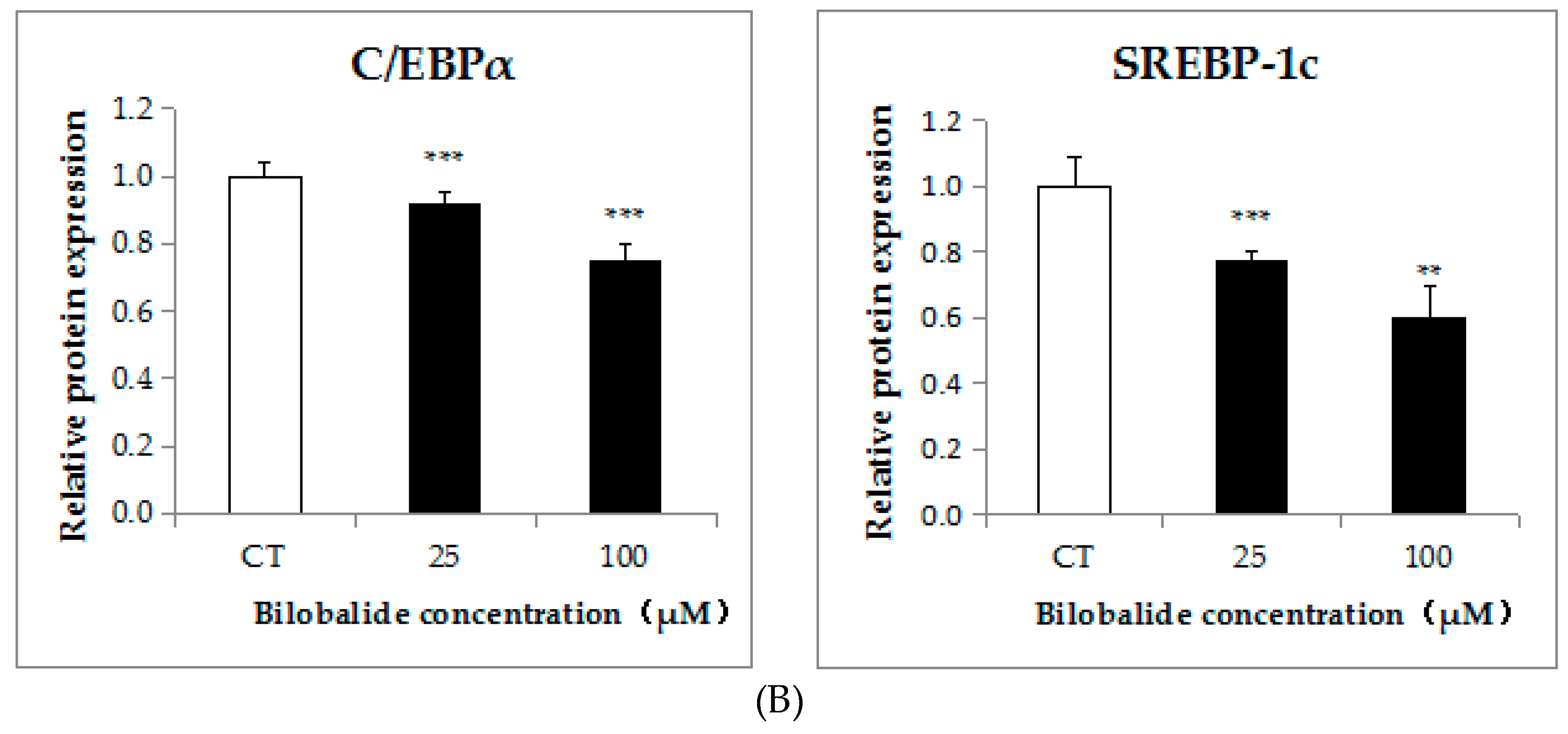
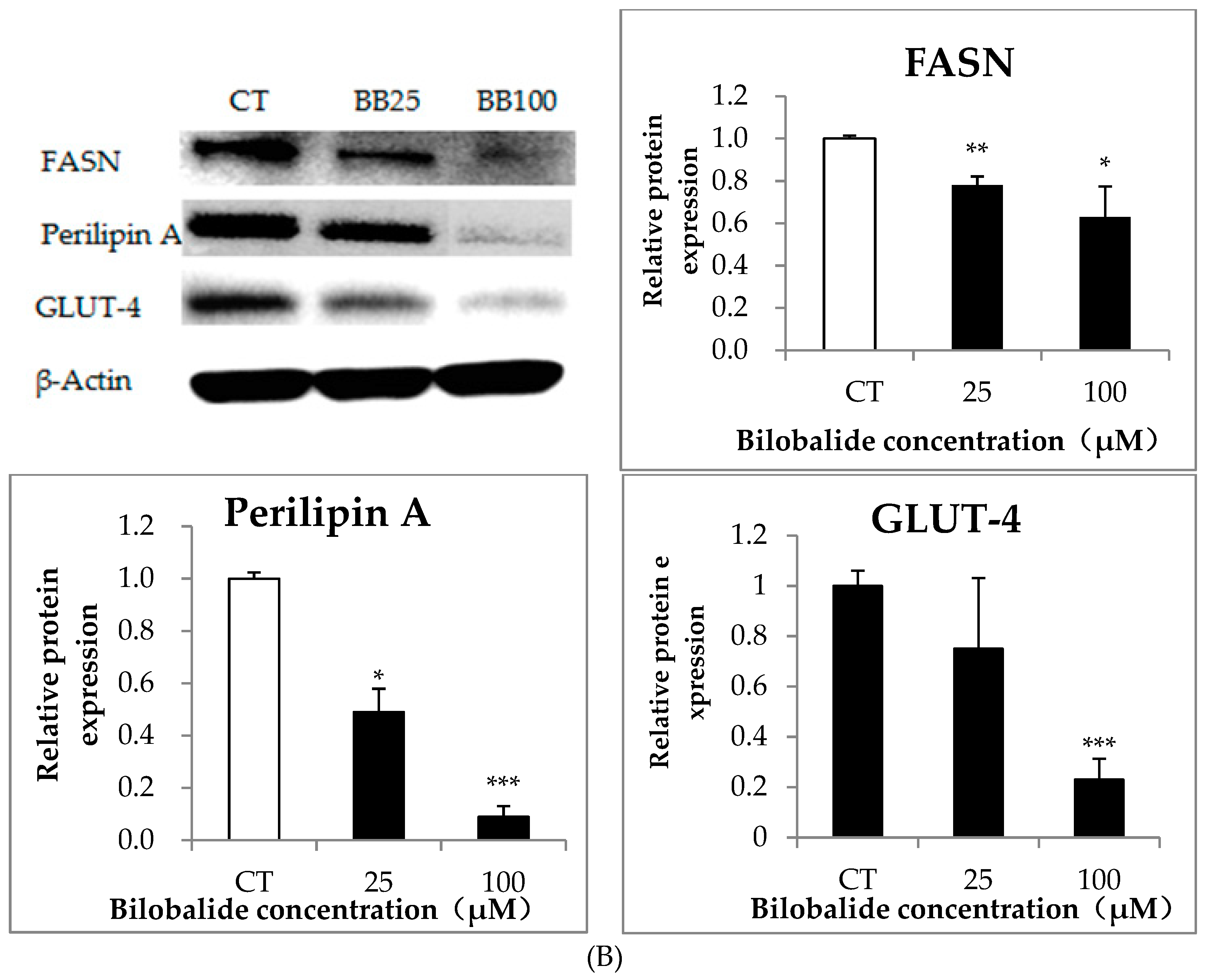
2.3. Bilobalide Downregulates the Expression of Adipogenic Transcription Factors
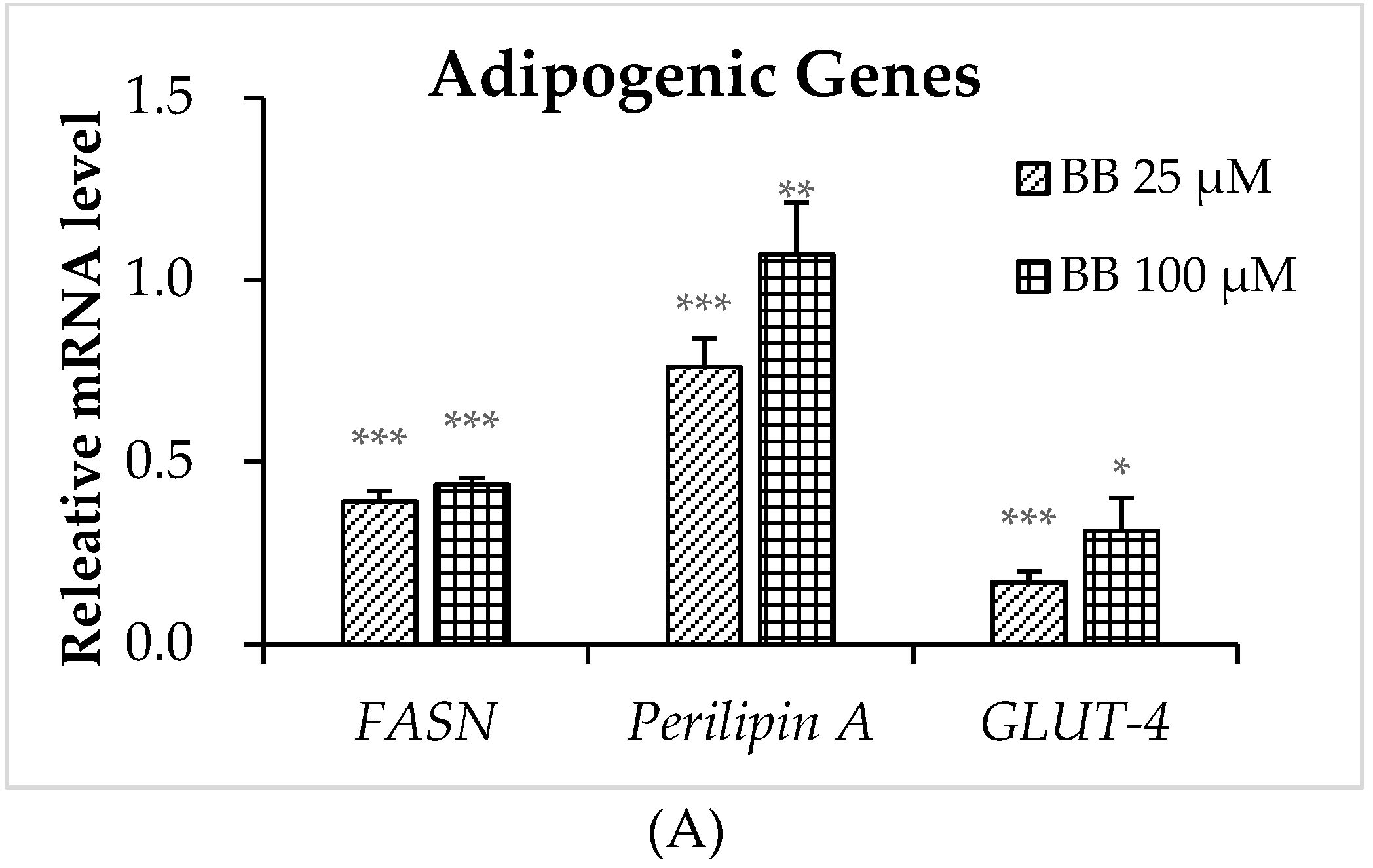
2.4. The Effect of Bilobalide on the Expression of Adipogenic Genes in Adipocytes
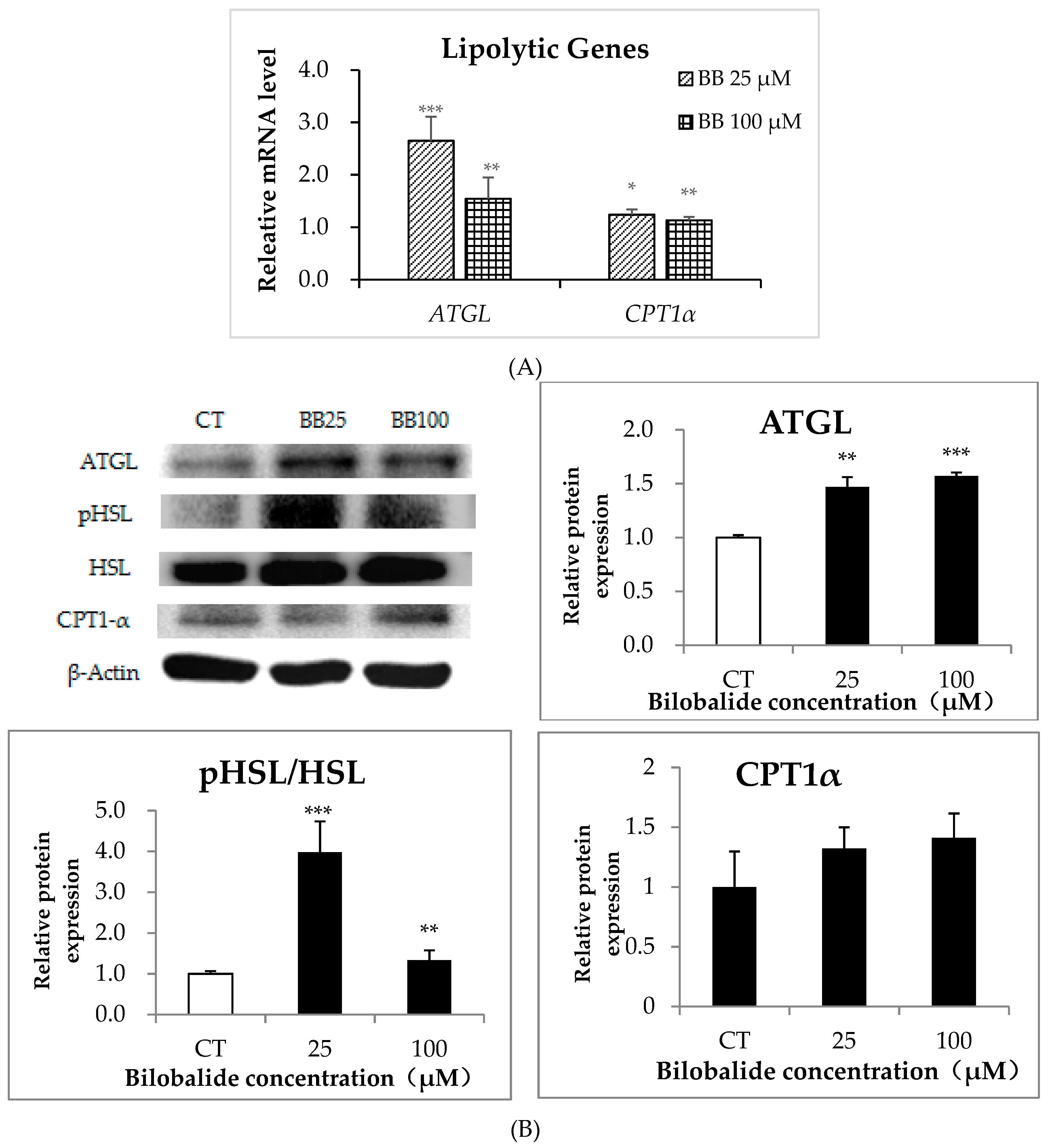
2.5. The Effect of Bilobalide on the Expression of Lipolytic Genes in 3T3-L1 Cells
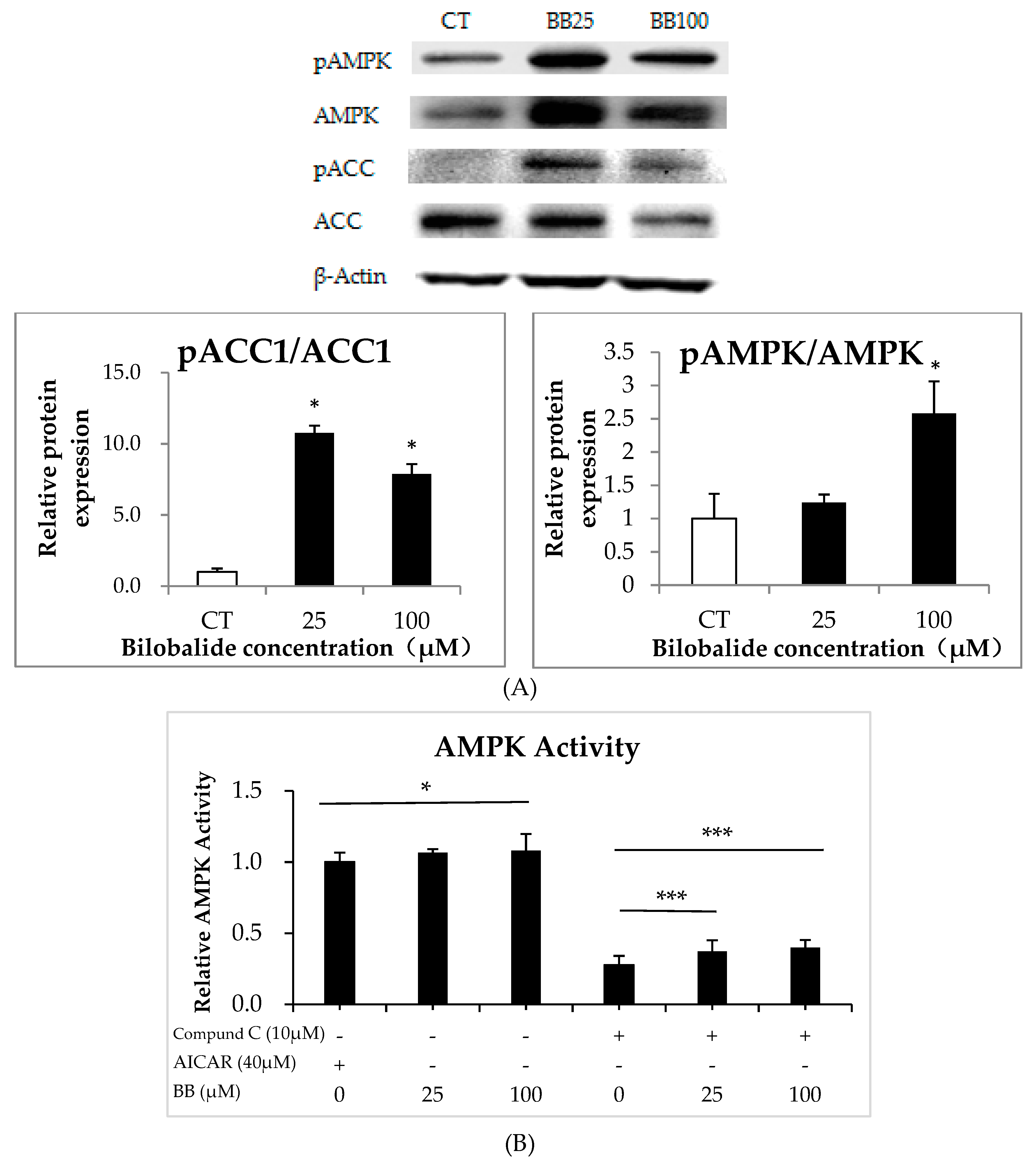
2.6. Bilobalide Activates AMPK Signaling
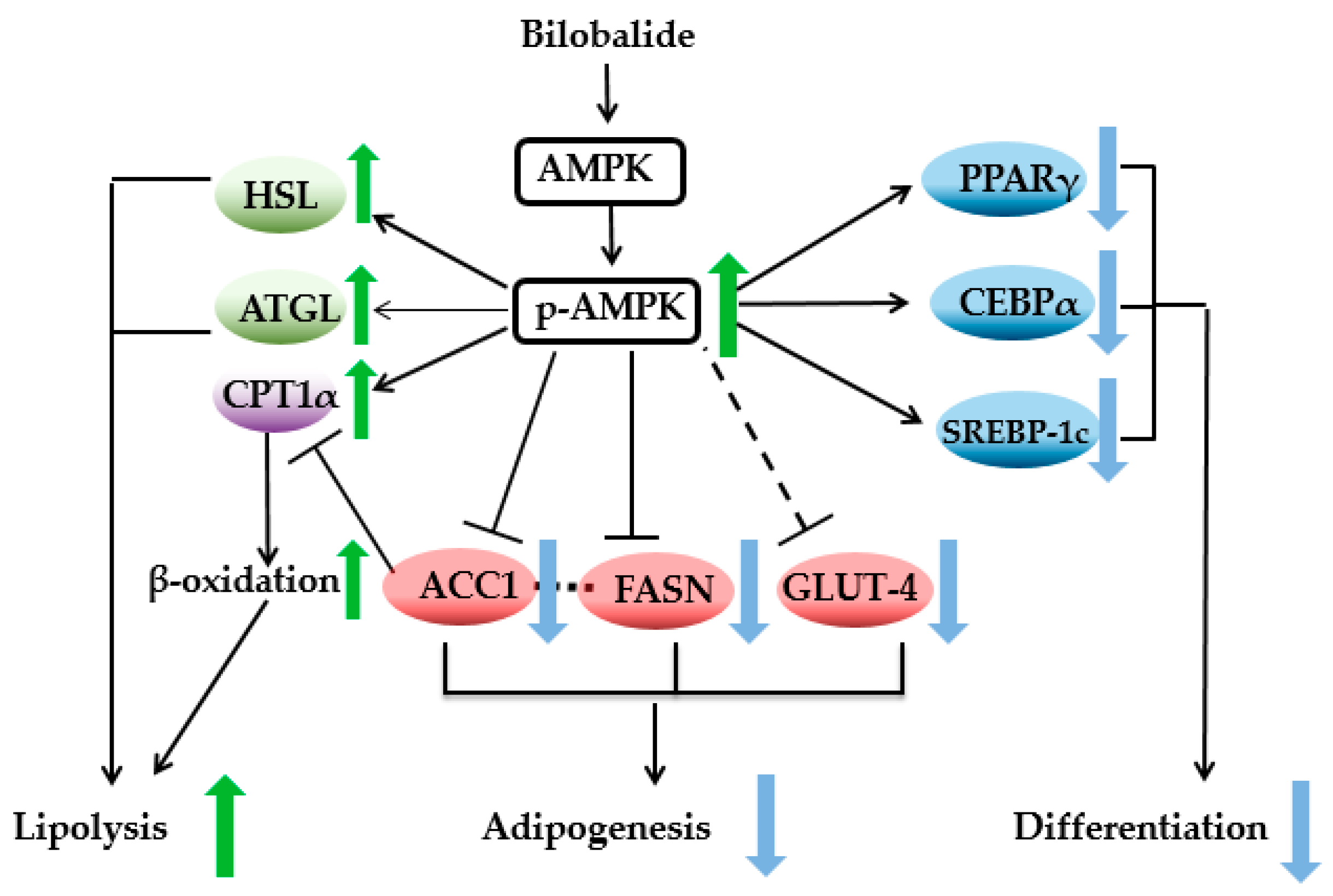
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Cell Culture
4.3. MTT Assay
4.4. ORO Staining
4.5. Quantification of the Expression of Adipogenic Genes in 3T3-L1 Cells
4.6. Western Blot
4.7. AMPK Activity Assay
4.8. Statistical Analysis
Author Contributions
Funding
Conflicts of Interest
References
- Palou, A.; Bonet, M.L. Challenges in obesity research. Nutr. Hosp. 2013, 28 (Suppl. 5), 144–153. [Google Scholar] [PubMed]
- Seidell, J.C.; Halberstadt, J. The global burden of obesity and the challenges of prevention. Ann. Nutr. Metab. 2015, 66 (Suppl. 2), 7–12. [Google Scholar] [CrossRef]
- Gonzalez-Muniesa, P.; Martinez-Gonzalez, M.A.; Hu, F.B.; Despres, J.P.; Matsuzawa, Y.; Loos, R.J.F.; Moreno, L.A.; Bray, G.A.; Martinez, J.A. Obesity. Nat. Rev. Dis. Primers 2017, 3, 17034. [Google Scholar] [CrossRef] [PubMed]
- Gregg, E.W.; Shaw, J.E. Global Health Effects of Overweight and Obesity. N. Engl. J. Med. 2017, 377, 80–81. [Google Scholar] [CrossRef] [PubMed]
- Ginsberg, H.N.; MacCallum, P.R. The obesity, metabolic syndrome, and type 2 diabetes mellitus pandemic: Part I. Increased cardiovascular disease risk and the importance of atherogenic dyslipidemia in persons with the metabolic syndrome and type 2 diabetes mellitus. J. Cardiometabolic Syndr. 2009, 4, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Garg, S.K.; Maurer, H.; Reed, K.; Selagamsetty, R. Diabetes and cancer: Two diseases with obesity as a common risk factor. Diabetes Obes. Metab. 2014, 16, 97–110. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.M.; Minson, C.T. Obesity and adipokines: Effects on sympathetic overactivity. J. Physiol. 2012, 590, 1787–1801. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Jiang, Y.; Ji, H.; Zhao, L.; Xiao, W.; Wang, Z.; Ding, G. The Synergistic Beneficial Effects of Ginkgo Flavonoid and Coriolus versicolor Polysaccharide for Memory Improvements in a Mouse Model of Dementia. Evid. Based Complementary Altern. Med. eCAM 2015, 2015, 128394. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, J.Q.; Fang, X.Y.; Ge, L.; Cao, F.L.; Zhao, L.G.; Wang, Z.Z.; Xiao, W. Antitumor, antioxidant and anti-inflammatory activities of kaempferol and its corresponding glycosides and the enzymatic preparation of kaempferol. PLoS ONE 2018, 13, e0197563. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekaran, K.; Mehrabian, Z.; Spinnewyn, B.; Drieu, K.; Fiskum, G. Neuroprotective effects of bilobalide, a component of the Ginkgo biloba extract (EGb 761), in gerbil global brain ischemia. Brain Res. 2001, 922, 282–292. [Google Scholar] [CrossRef]
- Kiewert, C.; Kumar, V.; Hildmann, O.; Rueda, M.; Hartmann, J.; Naik, R.S.; Klein, J. Role of GABAergic antagonism in the neuroprotective effects of bilobalide. Brain Res. 2007, 1128, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Guo, J.; Wang, J.; Zhang, L.; Pang, T.; Liao, H. Bilobalide induces neuronal differentiation of P19 embryonic carcinoma cells via activating Wnt/beta-catenin pathway. Cell. Mol. Neurobiol. 2014, 34, 913–923. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Wu, F.; Yew, D.T.; Xu, J.; Zhu, Y. Bilobalide prevents apoptosis through activation of the PI3K/Akt pathway in SH-SY5Y cells. Apoptosis Int. J. Program. Cell Death 2010, 15, 715–727. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.M.; Gu, S.S.; Mei, W.H.; Zhou, J.; Wang, Z.Z.; Xiao, W. Ginkgolides and bilobalide protect BV2 microglia cells against OGD/reoxygenation injury by inhibiting TLR2/4 signaling pathways. Cell Stress Chaperones 2016, 21, 1037–1053. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.S.; Kim, K.H.; Lee, I.S.; Park, J.Y.; Kim, Y.; Kim, K.S.; Jang, H.J. Ginkgolide A ameliorates non-alcoholic fatty liver diseases on high fat diet mice. Biomed. Pharmacother. 2017, 88, 625–634. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.C.; Chen, Y.L.; Liu, H.C.; Wu, S.J.; Liou, C.J. Ginkgolide C reduced oleic acid-induced lipid accumulation in HepG2 cells. Saudi Pharm. J. 2018, 26, 1178–1184. [Google Scholar] [CrossRef]
- Nguyen, M.T.; Csermely, P.; Soti, C. Hsp90 chaperones PPARgamma and regulates differentiation and survival of 3T3-L1 adipocytes. Cell Death Differ. 2013, 20, 1654–1663. [Google Scholar] [CrossRef] [PubMed]
- Shao, H.Y.; Hsu, H.Y.; Wu, K.S.; Hee, S.W.; Chuang, L.M.; Yeh, J.I. Prolonged induction activates Cebpalpha independent adipogenesis in NIH/3T3 cells. PLoS ONE 2013, 8, e51459. [Google Scholar]
- Ma, C.; Li, G.; He, Y.; Xu, B.; Mi, X.; Wang, H.; Wang, Z. Pronuciferine and nuciferine inhibit lipogenesis in 3T3-L1 adipocytes by activating the AMPK signaling pathway. Life Sci. 2015, 136, 120–125. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhao, X.; Feng, X.; Liu, X.; Deng, C.; Hu, C.H. Berberine Alleviates Olanzapine-Induced Adipogenesis via the AMPKalpha-SREBP Pathway in 3T3-L1 Cells. Int. J. Mol. Sci. 2016, 17, 1865. [Google Scholar] [CrossRef]
- Kim, E.J.; Lee, D.H.; Kim, H.J.; Lee, S.J.; Ban, J.O.; Cho, M.C.; Jeong, H.S.; Yang, Y.; Hong, J.T.; Yoon, D.Y. Thiacremonone, a sulfur compound isolated from garlic, attenuates lipid accumulation partially mediated via AMPK activation in 3T3-L1 adipocytes. J. Nutr. Biochem. 2012, 23, 1552–1558. [Google Scholar] [CrossRef] [PubMed]
- Marsh, B.J.; Alm, R.A.; McIntosh, S.R.; James, D.E. Molecular regulation of GLUT-4 targeting in 3T3-L1 adipocytes. J. Cell Biol. 1995, 130, 1081–1091. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.; Yijun, L.; Jingtao, D.; Changyu, P.; Wenhua, Y.; Baoan, W.; Fangling, M.; Xianling, W.; Guoqing, Y.; Yiming, M.; et al. Effects of telmisartan on lipid metabolisms and proinflammatory factors secretion of differentiated 3T3-L1 adipocytes. J. Renin Angiotensin Aldosterone Syst. JRAAS 2015, 16, 1061–1068. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.W.; Lee, J.; Park, S.E.; Rhee, E.J.; Park, C.Y.; Oh, K.W.; Park, S.W.; Lee, W.Y. Activation of AMP-Activated Protein Kinase Attenuates Tumor Necrosis Factor-alpha-Induced Lipolysis via Protection of Perilipin in 3T3-L1 Adipocytes. Endocrinol. Metab. 2014, 29, 553–560. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.O.; Sakchaisri, K.; Asami, Y.; Ryoo, I.J.; Choo, S.J.; Yoo, I.D.; Soung, N.K.; Kim, Y.S.; Jang, J.H.; Kim, B.Y.; et al. Illudins C2 and C3 stimulate lipolysis in 3T3-L1 adipocytes and suppress adipogenesis in 3T3-L1 preadipocytes. J. Nat. Prod. 2014, 77, 744–750. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.; Lee, H.; Kim, S.; Ha, T. Curcumin-induced suppression of adipogenic differentiation is accompanied by activation of Wnt/beta-catenin signaling. Am. J. Physiol. Cell Physiol. 2010, 298, C1510–C1516. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.X.; Cheng, X.Y.; Wang, Y.; Yin, W. Toosendanin inhibits adipogenesis by activating Wnt/beta-catenin signaling. Sci. Rep. 2018, 8, 4626. [Google Scholar] [CrossRef] [PubMed]
- Choe, W.K.; Kang, B.T.; Kim, S.O. Water-extracted plum (Prunus salicina L. cv. Soldam) attenuates adipogenesis in murine 3T3-L1 adipocyte cells through the PI3K/Akt signaling pathway. Exp. Ther. Med. 2018, 15, 1608–1615. [Google Scholar] [PubMed]
- Wu, Z.; Zhang, J.D.; Gu, X.; Zhang, X.X.; Shi, S.M.; Liu, C. Effects of the extract of Ginkgo biloba on the differentiation of bone marrow mesenchymal stem cells in vitro. Am. J. Transl. Res. 2016, 8, 3032–3040. [Google Scholar] [PubMed]
- Banin, R.M.; Hirata, B.K.S.; Andrade, I.S.; Zemdegs, J.C.S.; Clemente, A.P.G.; Dornellas, A.P.S.; Boldarine, V.T.; Estadella, D.; Albuquerque, K.T.; Oyama, L.M.; et al. Beneficial effects of Ginkgo biloba extract on insulin signaling cascade, dyslipidemia, and body adiposity of diet-induced obese rats. Braz. J. Med. Biol. Res. 2014, 47, 780–788. [Google Scholar] [CrossRef]
- Wei, T.; Xiong, F.F.; Wang, S.D.; Wang, K.; Zhang, Y.Y.; Zhang, Q.H. Flavonoid ingredients of Ginkgo biloba leaf extract regulate lipid metabolism through Sp1-mediated carnitine palmitoyltranferase 1A up-regulation. J. Biomed. Sci. 2014, 21, 87. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.S.; Kim, Y. Effects of Isorhamnetin on Adipocyte Mitochondrial Biogenesis and AMPK Activation. Molecules 2018, 23, 1853. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.J.; Choi, H.S.; Seo, M.J.; Jeon, H.J.; Kim, K.J.; Lee, B.Y. Kaempferol suppresses lipid accumulation by inhibiting early adipogenesis in 3T3-L1 cells and zebrafish. Food Funct. 2015, 6, 2824–2833. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.L.; Hu, J.J.; Zhao, W.W.; Gao, X.J.; Jiang, C.H.; Liu, K.; Liu, B.L.; Huang, F. Quercetin differently regulates insulin-mediated glucose transporter 4 translocation under basal and inflammatory conditions in adipocytes. Mol. Nutr. Food Res. 2014, 58, 931–941. [Google Scholar] [CrossRef] [PubMed]
- Liou, C.J.; Lai, X.Y.; Chen, Y.L.; Wang, C.L.; Wei, C.H.; Huang, W.C. Ginkgolide C Suppresses Adipogenesis in 3T3-L1 Adipocytes via the AMPK Signaling Pathway. Evid. Based Complementary Altern. Med. eCAM 2015, 2015, 298635. [Google Scholar] [CrossRef] [PubMed]
- Mihaylova, M.M.; Shaw, R.J. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat. Cell Biol. 2011, 13, 1016–1023. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.B.; Zhou, G.C.; Li, C. AMPK: An Emerging Drug Target for Diabetes and the Metabolic Syndrome. Cell Metab. 2009, 9, 407–416. [Google Scholar] [CrossRef] [PubMed]
- Davies, S.P.; Carling, D.; Munday, M.R.; Hardie, D.G. Diurnal rhythm of phosphorylation of rat liver acetyl-CoA carboxylase by the AMP-activated protein kinase, demonstrated using freeze-clamping. Effects of high fat diets. Eur. J. Biochem. 1992, 203, 615–623. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Peng, Y.; Duan, W.; Tian, Y.; Zhang, J.; Hu, T.; Cai, Y.; Feng, Y.; Li, G. Aspirin regulates hepatocellular lipid metabolism by activating AMPK signaling pathway. J. Toxicol. Sci. 2015, 40, 127–136. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Young, S.G.; Zechner, R. Biochemistry and pathophysiology of intravascular and intracellular lipolysis. Genes Dev. 2013, 27, 459–484. [Google Scholar] [CrossRef] [PubMed]
- Fruhbeck, G.; Mendez-Gimenez, L.; Fernandez-Formoso, J.A.; Fernandez, S.; Rodriguez, A. Regulation of adipocyte lipolysis. Nutr. Res. Rev. 2014, 27, 63–93. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.L.; Zhou, F.; Jiang, H.T.; Wang, Z.J.; Hua, C.; Zhang, Y.S. Chicory (Cichorium intybus L.) polysaccharides attenuate high-fat diet induced non-alcoholic fatty liver disease via AMPK activation. Int. J. Biol. Macromol. 2018, 118, 886–895. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.Y.; Zhang, M.L.; Ma, Y.Q.; Lu, J.X.; Pan, J.M.; Pan, P.; Chen, H.B.; Jia, W.P. 5-ALA ameliorates hepatic steatosis through AMPK signaling pathway. J. Mol. Endocrinol. 2017, 59, 121–128. [Google Scholar] [CrossRef]
- Porstmann, T.; Santos, C.R.; Griffiths, B.; Cully, M.; Wu, M.; Leevers, S.; Griffiths, J.R.; Chung, Y.L.; Schulze, A. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 2008, 8, 224–236. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xu, S.; Mihaylova, M.M.; Zheng, B.; Hou, X.; Jiang, B.; Park, O.; Luo, Z.; Lefai, E.; Shyy, J.Y.; et al. AMPK phosphorylates and inhibits SREBP activity to attenuate hepatic steatosis and atherosclerosis in diet-induced insulin-resistant mice. Cell Metab. 2011, 13, 376–388. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.K.; Obiang-Obounou, B.W.; Lee, K.B.; Choi, J.S.; Jang, B.C. AZD1208, a pan-Pim kinase inhibitor, inhibits adipogenesis and induces lipolysis in 3T3-L1 adipocytes. J. Cell. Mol. Med. 2018, 22, 2488–2497. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, S.D.; Wang, P.; Guo, N.; Wang, W.; Yao, L.P.; Yang, Q.; Efferth, T.; Jiao, J.; Fu, Y.J. Pinolenic acid ameliorates oleic acid-induced lipogenesis and oxidative stress via AMPK/SIRT1 signaling pathway in HepG2 cells. Eur. J. Pharmacol. 2019, 861, 172618. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.Y.; Shi, Z.H.; Ma, W.; Tao, D.Q.; Liu, S.; Chen, L.; Zhou, X.L. Effect of Fuzi Lizhong decoction in reducing liver injury of rats with non-alcoholic fatty liver via activating AMPK and suppressing NF-kappaBp65 pathway. China J. Chin. Mater. Med. 2018, 43, 3176–3183. [Google Scholar]
- Moraes-Vieira, P.M.; Saghatelian, A.; Kahn, B.B. GLUT4 Expression in Adipocytes Regulates De Novo Lipogenesis and Levels of a Novel Class of Lipids with Antidiabetic and Anti-inflammatory Effects. Diabetes 2016, 65, 1808–1815. [Google Scholar] [CrossRef] [PubMed]
- Les, F.; Arbones-Mainar, J.M.; Valero, M.S.; Lopez, V. Pomegranate polypheno and urolithin A inhibit alpha-glucosidase, dipeptidyl peptidase-4, lipase, triglyceride accumulation and adipogenesis related genes in 3T3-L1 adipocyte-like cells. J. Ethnopharmacol. 2018, 220, 67–74. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |

| Name | GenBank No. | Primer Sequence (5’– 3’) |
|---|---|---|
| PPARγ | NM_001308354.1 | F: AGGCCGAGAAGGAGAAGCTGTTG R: TGGCCACCTCTTTGCTCTGCTC |
| C/EBPα | NM_001308354.1 | F: GCCCCCGTGAGAAAAATGAAG R: GAGGTGCGAAAAGCAAGGGA |
| FASN | NM_007988.3 | F: ATTCGGTGTATCCTGCTGTC R: GCTTGTCCTGCTCTAACTGG |
| SREBP-1c | NM_001278601 | F: TGGACTACTAGTGTTGGCCTGCTT R: ATCCAGGTCAGCTTGTTTGCGATG |
| GLUT-4 | NM_009204 | F: AGCCTCTGATCATCGCAGTG R: ACCGAGACCAACGTGAAGAC |
| ATGL | NM_001163689 | F: GGTTAGAGTTGCTCAGCCGT R: ACATGAGGAGCGGATGTGTG |
| CPT1α | NM_013495 | F: GGGCTTGGTAGTCAAAGGCT R: TGCCTGTGTCAGTATGCCTG |
| Perilipin A | NM_001113471.1 | F: CACTCTCTGGCCATGTGGA R: AGAGGCTGCCAGGTTGTG |
| 18S rRNA | NR_003278.3 | F: ATGCGGCGGCGTTATTCC R: CTGTCAATCCTGTCCGTGTCC |
| GAPDH | XM_029476871.1 | F: CAGGTTGTCTCCTGCGACTT R: TATGGGGGTCTGGGATGGAA |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bu, S.; Yuan, C.Y.; Xue, Q.; Chen, Y.; Cao, F. Bilobalide Suppresses Adipogenesis in 3T3-L1 Adipocytes via the AMPK Signaling Pathway. Molecules 2019, 24, 3503. https://doi.org/10.3390/molecules24193503
Bu S, Yuan CY, Xue Q, Chen Y, Cao F. Bilobalide Suppresses Adipogenesis in 3T3-L1 Adipocytes via the AMPK Signaling Pathway. Molecules. 2019; 24(19):3503. https://doi.org/10.3390/molecules24193503
Chicago/Turabian StyleBu, Su, Chun Ying Yuan, Quan Xue, Ying Chen, and Fuliang Cao. 2019. "Bilobalide Suppresses Adipogenesis in 3T3-L1 Adipocytes via the AMPK Signaling Pathway" Molecules 24, no. 19: 3503. https://doi.org/10.3390/molecules24193503
APA StyleBu, S., Yuan, C. Y., Xue, Q., Chen, Y., & Cao, F. (2019). Bilobalide Suppresses Adipogenesis in 3T3-L1 Adipocytes via the AMPK Signaling Pathway. Molecules, 24(19), 3503. https://doi.org/10.3390/molecules24193503





