Trilobatin, a Novel SGLT1/2 Inhibitor, Selectively Induces the Proliferation of Human Hepatoblastoma Cells
Abstract
1. Introduction
2. Results
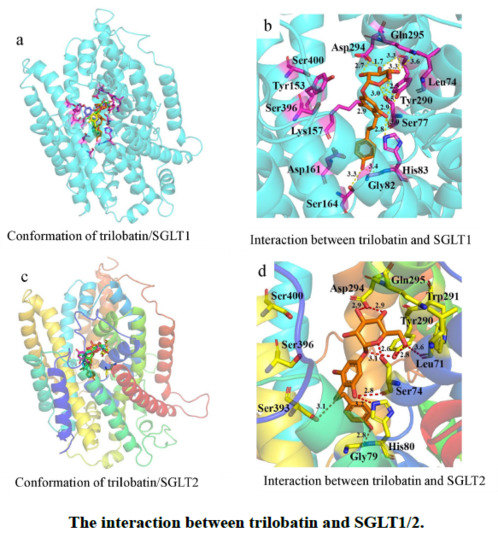
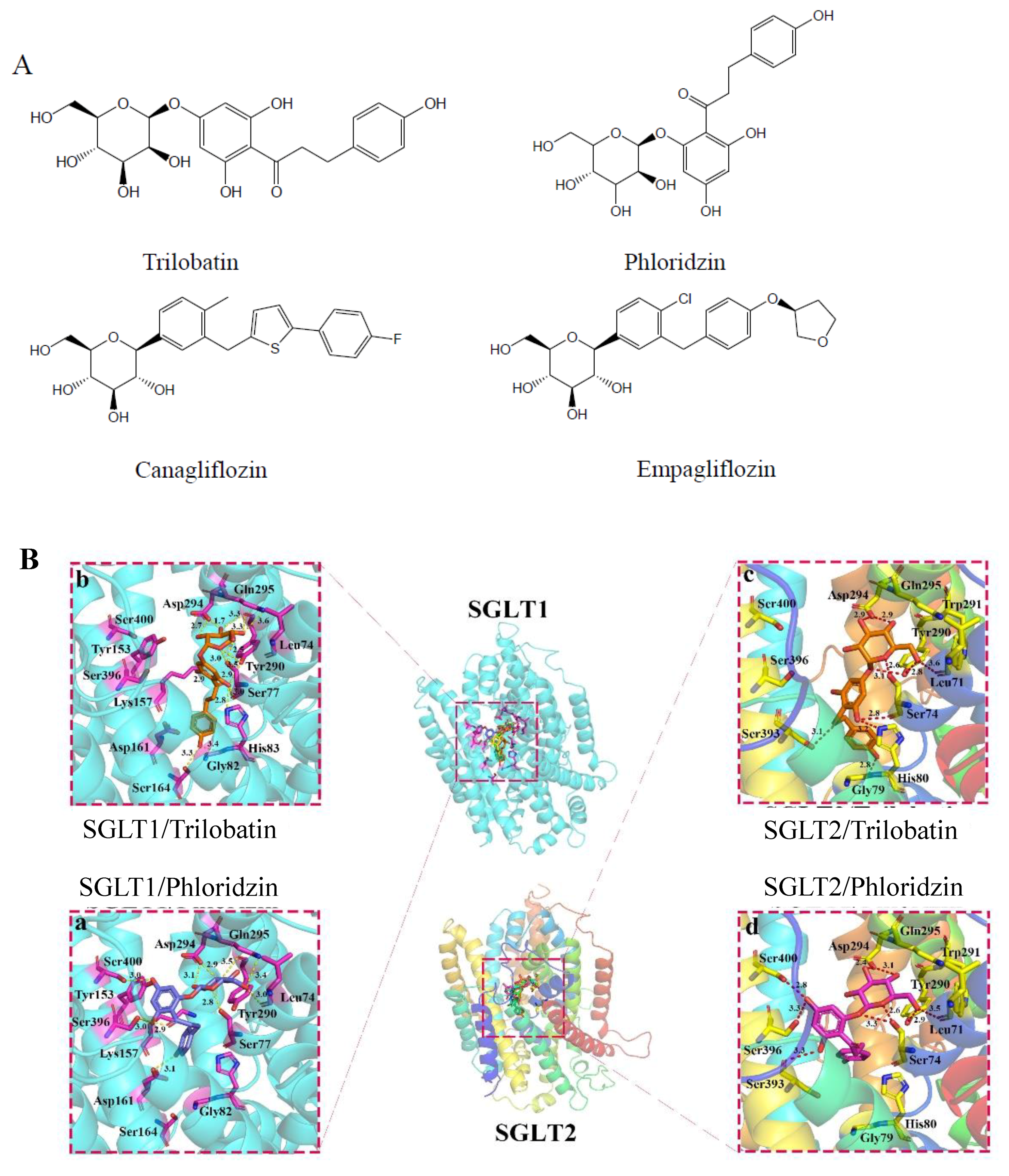
2.1. Molecular Docking of SGLT1/2 Inhibitors
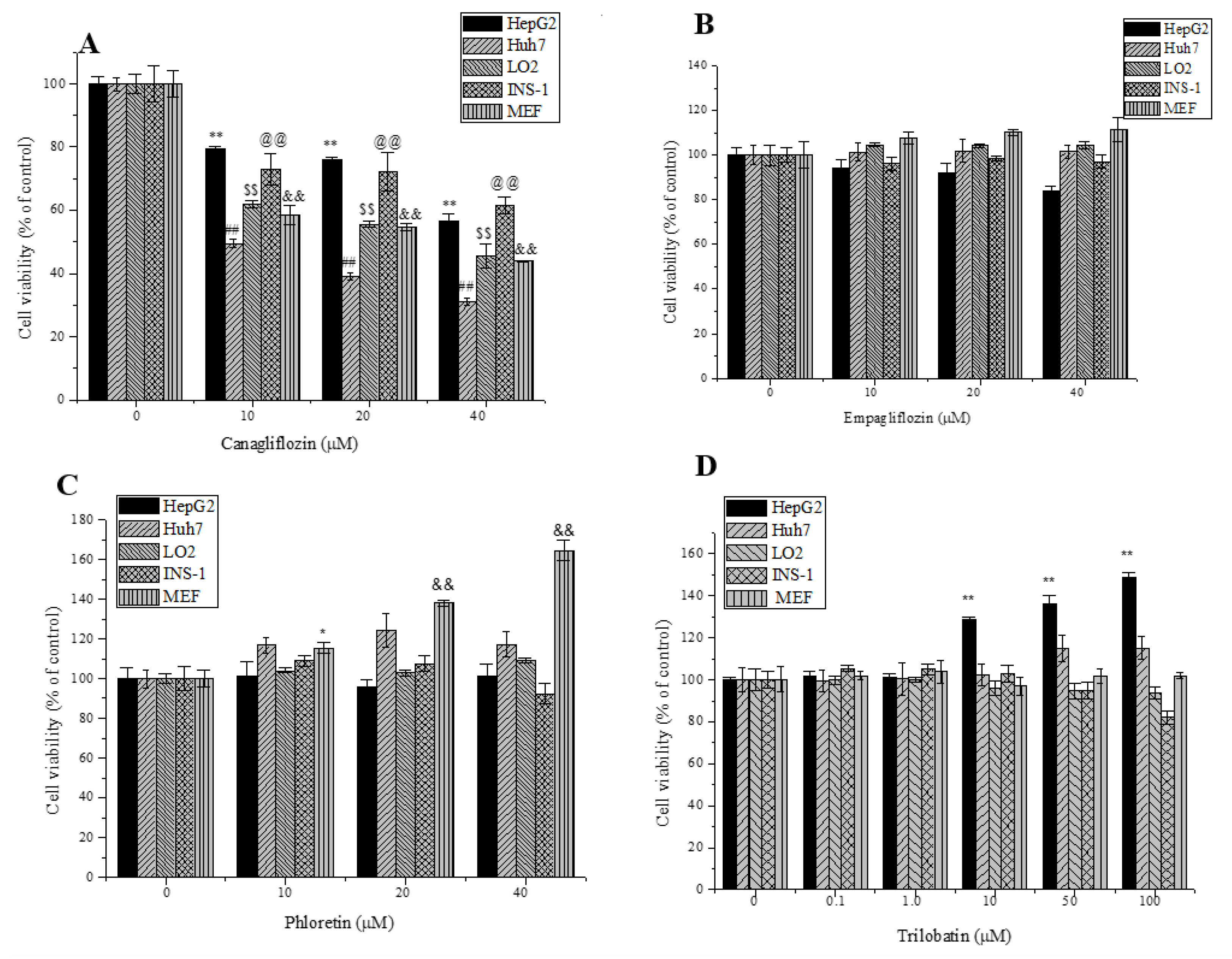
2.2. Effects of Trilobatin, Phloridzin, Canagliflozin, and Empagliflozin on the Cell Proliferation
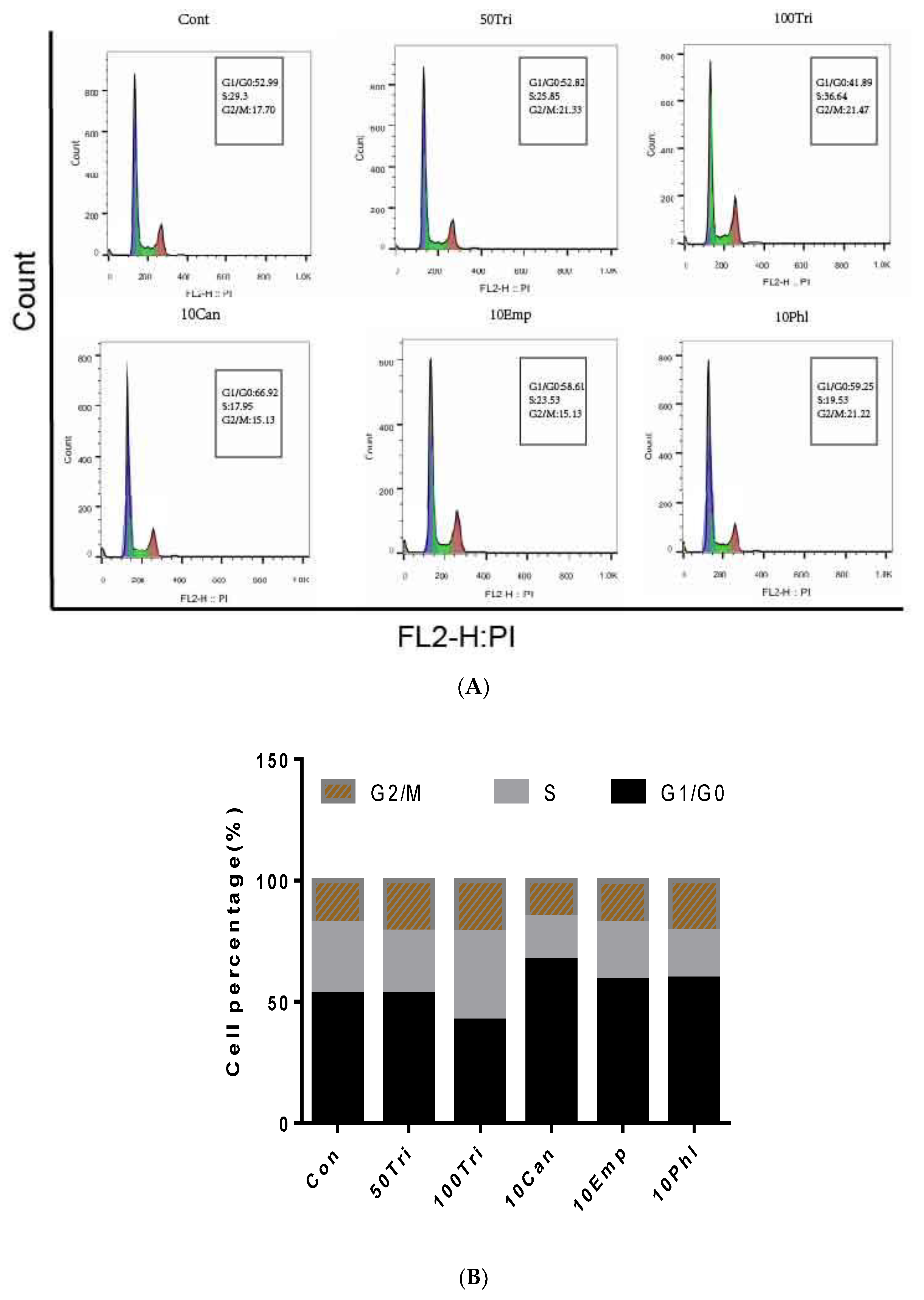
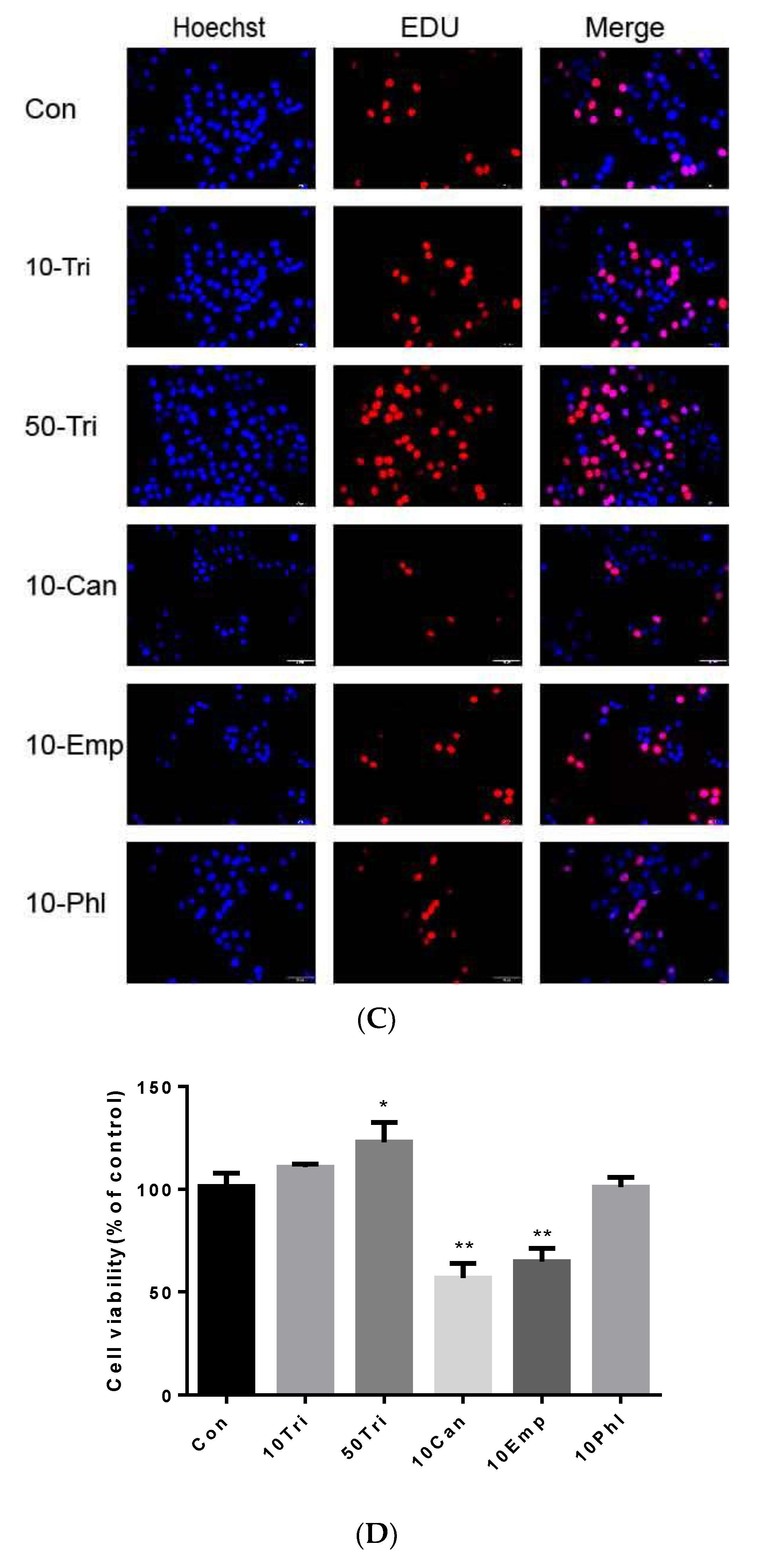
2.3. Effects of Trilobatin, Phloridzin, Canagliflozin, and Empagliflozin on the Cell Cycle and DNA Replication in HepG2 cells
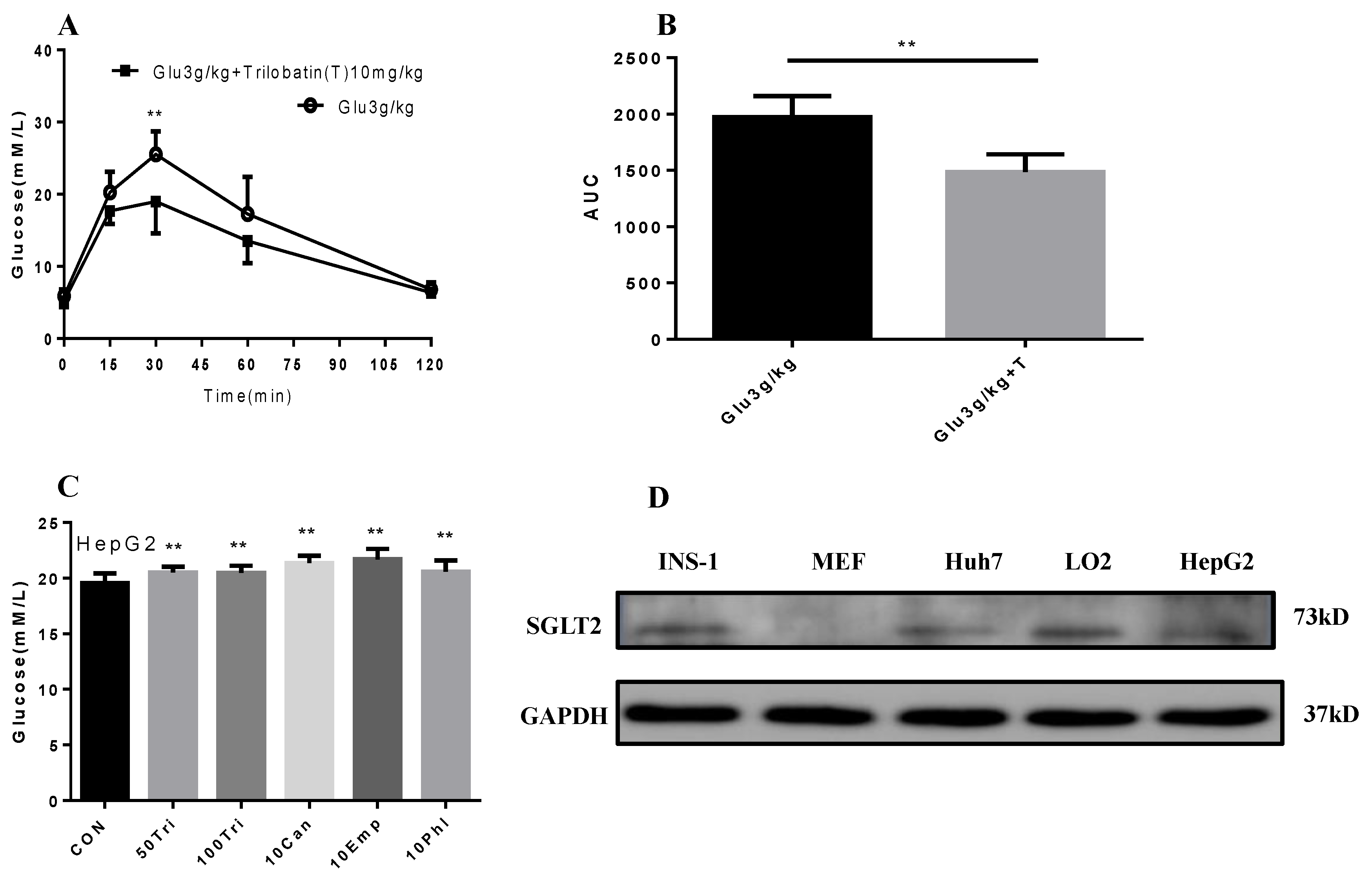
2.4. Trilobatin Inhibits Intake of Glucose in Vivo and in Vitro
2.5. Effect of Trilobatin on the Expression of HBXIP and HNF4α in HepG2 Cells
3. Materials and Methods
3.1. Materials
3.2. Cell Culture
3.3. Molecular Docking of SGLT1/2 Inhibitors
3.4. Glucose Uptake in Mice
3.5. MTT Assay
3.6. EDU Staining
3.7. Cell Cycle Assay
3.8. RNA Extraction and Quantitative Real-Time PCR
3.9. Western Blot
3.10. Statistical Analysis
4. Discussion
5. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Ganapathy-Kanniappan, S.; Geschwind, J.F. Tumor glycolysis as a target for cancer therapy: Progress and prospects. Mol. Cancer 2013, 12, 152. [Google Scholar] [CrossRef] [PubMed]
- Teicher, B.A.; Linehan, W.M.; Helman, L.J. Targeting cancer metabolism. Clin. Cancer Res. 2012, 18, 5537–5545. [Google Scholar] [CrossRef] [PubMed]
- Koepsell, H. The Na(+)-d-glucose cotransporters SGLT1 and SGLT2 are targets for the treatment of diabetes and cancer. Pharmacol. Ther. 2017, 170, 148–165. [Google Scholar] [CrossRef] [PubMed]
- Scafoglio, C.; Hirayama, B.A.; Kepe, V.; Liu, J.; Ghezzi, C.; Satyamurthy, N.; Moatamed, N.A.; Huang, J.; Koepsell, H.; Barrio, J.R.; et al. Functional expression of sodium-glucose transporters in cancer. Proc. Natl. Acad. Sci. USA 2015, 112, E4111–E4119. [Google Scholar] [CrossRef] [PubMed]
- Ehrenkranz, J.R.; Lewis, N.G.; Kahn, C.R.; Roth, J. Phlorizin: A review. Diabetes Metab. Res. Rev. 2005, 21, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Najafian, M.; Jahromi, M.Z.; Nowroznejhad, M.J.; Khajeaian, P.; Kargar, M.M.; Sadeghi, M.; Arasteh, A. Phloridzin reduces blood glucose levels and improves lipids metabolism in streptozotocin-induced diabetic rats. Mol. Biol. Rep. 2012, 39, 5299–5306. [Google Scholar] [CrossRef] [PubMed]
- Nelson, J.A.; Falk, R.E. The efficacy of phloridzin and phloretin on tumor cell growth. Anticancer Res. 1993, 13, 2287–2292. [Google Scholar] [PubMed]
- Wang, J.; Huang, Y.; Li, K.; Chen, Y.; Vanegas, D.; McLamore, E.S.; Shen, Y. Leaf Extract from Lithocarpus polystachyus Rehd. Promote Glycogen Synthesis in T2DM Mice. PLoS ONE 2016, 11, e0166557. [Google Scholar] [CrossRef]
- Sun, Y.; Li, W.; Liu, Z. Preparative isolation, quantification and antioxidant activity of dihydrochalcones from Sweet Tea (Lithocarpus polystachyus Rehd.). J. Chromatogr. B 2015, 1002, 372–378. [Google Scholar] [CrossRef]
- Fan, X.; Zhang, Y.; Dong, H.; Wang, B.; Ji, H.; Liu, X. Trilobatin attenuates the LPS-mediated inflammatory response by suppressing the NF-kappaB signaling pathway. Food Chem. 2015, 166, 609–615. [Google Scholar] [CrossRef]
- Qin, X.; Xing, Y.F.; Zhou, Z.; Yao, Y. Dihydrochalcone Compounds Isolated from Crabapple Leaves Showed Anticancer Effects on Human Cancer Cell Lines. Molecules 2015, 20, 21193–21203. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, A.; Choe, S.; Chaptal, V.; Rosenberg, J.M.; Wright, E.M.; Grabe, M.; Abramson, J. The mechanism of sodium and substrate release from the binding pocket of vSGLT. Nature 2010, 468, 988. [Google Scholar] [CrossRef] [PubMed]
- Perez, M.; Praena-Fernandez, J.M.; Felipe-Abrio, B.; Lopez-Garcia, M.A.; Lucena-Cacace, A.; Garcia, A.; Lleonart, M.; Roncador, G.; Marin, J.J.; Carnero, A. MAP17 and SGLT1 protein expression levels as prognostic markers for cervical tumor patient survival. PLoS ONE 2013, 8, e56169. [Google Scholar] [CrossRef] [PubMed]
- Kaji, K.; Nishimura, N.; Seki, K.; Sato, S.; Saikawa, S.; Nakanishi, K.; Furukawa, M.; Kawaratani, H.; Kitade, M.; Moriya, K.; et al. Sodium glucose cotransporter 2 inhibitor canagliflozin attenuates liver cancer cell growth and angiogenic activity by inhibiting glucose uptake. Int J. Cancer 2018, 142, 1712–1722. [Google Scholar] [CrossRef] [PubMed]
- Yamakoshi, H.; Dodo, K.; Okada, M.; Ando, J.; Palonpon, A.; Fujita, K.; Kawata, S.; Sodeoka, M. Imaging of EdU, an alkyne-tagged cell proliferation probe, by Raman microscopy. J. Am. Chem. Soc. 2011, 133, 6102–6105. [Google Scholar] [CrossRef] [PubMed]
- Diermeier-Daucher, S.; Clarke, S.T.; Hill, D.; Vollmann-Zwerenz, A.; Bradford, J.A.; Brockhoff, G. Cell type specific applicability of 5-ethynyl-2′-deoxyuridine (EdU) for dynamic proliferation assessment in flow cytometry. Cytometry A 2009, 75, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Salic, A.; Mitchison, T.J. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc. Natl. Acad. Sci. USA 2008, 105, 2415–2420. [Google Scholar] [CrossRef] [PubMed]
- Okada, M.; Shi, Y.B. Cell Proliferation Analysis during Xenopus Metamorphosis: Using 5-Ethynyl-2-Deoxyuridine (EdU) to Stain Proliferating Intestinal Cells. Cold Spring Harb. Protoc. 2017, 2017. [Google Scholar] [CrossRef]
- Limsirichaikul, S.; Niimi, A.; Fawcett, H.; Lehmann, A.; Yamashita, S.; Ogi, T. A rapid non-radioactive technique for measurement of repair synthesis in primary human fibroblasts by incorporation of ethynyl deoxyuridine (EdU). Nucleic Acids Res. 2009, 37, e31. [Google Scholar] [CrossRef]
- Wang, F.Z.; Sha, L.; Zhang, W.Y.; Wu, L.Y.; Qiao, L.; Li, N.; Zhang, X.D.; Ye, L.H. Involvement of hepatitis B X-interacting protein (HBXIP) in proliferation regulation of cells. Acta Pharmacol. Sin. 2007, 28, 431–438. [Google Scholar] [CrossRef][Green Version]
- Wang, F.Z.; Fei, H.R.; Lian, L.H.; Wang, J.M.; Qiu, Y.Y. Hepatitis B x-interacting protein induces HepG2 cell proliferation through activation of the phosphatidylinositol 3-kinase/Akt pathway. Exp. Biol. Med. (Maywood) 2011, 236, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Wang, Z.H.; Cheng, S.B.; Li, H.K.; Chan, H.B.; Chew, E.C. The effect of arsenic trioxide on the expression of Hsc and HNF4 in nuclear matrix proteins in HepG2 cells. Anticancer Res. 2001, 21, 2553–2559. [Google Scholar]
- Eswar, N.; Webb, B.; Marti-Renom, M.A.; Madhusudhan, M.S.; Eramian, D.; Shen, M.Y.; Pieper, U.; Sali, A. Comparative protein structure modeling using Modeller. Curr. Protoc. Bioinform. 2006, 5, 5–6. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.N. Scoring noncovalent protein-ligand interactions: A continuous differentiable function tuned to compute binding affinities. J. Comput. Aided Mol. Des. 1996, 10, 427–440. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Kochanek, S.; Close, D.; Wang, L.; Srinivasan, A.; Almehizia, A.A.; Iyer, P.; Xie, X.Q.; Johnston, P.A.; Gold, B. Design and activity of AP endonuclease-1 inhibitors. J. Chem. Biol. 2015, 8, 79–93. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Pearce, L.V.; Xu, X.; Yang, X.; Yang, P.; Blumberg, P.M.; Xie, X.Q. Structural insight into tetrameric hTRPV1 from homology modeling, molecular docking, molecular dynamics simulation, virtual screening, and bioassay validations. J. Chem. Inf. Model. 2015, 55, 572–588. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lin, W.; Wu, N.; He, X.; Wang, J.; Feng, Z.; Xie, X.Q. An insight into paracetamol and its metabolites using molecular docking and molecular dynamics simulation. J. Mol. Model. 2018, 24, 243. [Google Scholar] [CrossRef]
- Wang, Y.Q.; Lin, W.W.; Wu, N.; Wang, S.Y.; Chen, M.Z.; Lin, Z.H.; Xie, X.Q.; Feng, Z.W. Structural insight into the serotonin (5-HT) receptor family by molecular docking, molecular dynamics simulation and systems pharmacology analysis. Acta Pharmacol. Sin. 2019, 40, 1138–1156. [Google Scholar] [CrossRef]
- Scafoglio, C.R.; Villegas, B.; Abdelhady, G.; Bailey, S.T.; Liu, J.; Shirali, A.S.; Wallace, W.D.; Magyar, C.E.; Grogan, T.R.; Elashoff, D.; et al. Sodium-glucose transporter 2 is a diagnostic and therapeutic target for early-stage lung adenocarcinoma. Sci. Transl. Med. 2018, 10, eaat5933. [Google Scholar] [CrossRef]
- Szablewski, L. Expression of glucose transporters in cancers. Biochim. Biophys. Acta 2013, 1835, 164–169. [Google Scholar] [CrossRef]
- Ren, W.; Qiao, Z.; Wang, H.; Zhu, L.; Zhang, L. Flavonoids: Promising anticancer agents. Med. Res. Rev. 2003, 23, 519–534. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, N.; Oguri, T.; Isobe, T.; Fujitaka, K.; Kohno, N. SGLT gene expression in primary lung cancers and their metastatic lesions. Jpn. J. Cancer Res. 2001, 92, 874–879. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds trilobatin and phloridzin are available from the authors. |

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Liu, M.; Yin, F.; Wang, Y.; Li, X.; Wu, Y.; Ye, C.; Liu, J. Trilobatin, a Novel SGLT1/2 Inhibitor, Selectively Induces the Proliferation of Human Hepatoblastoma Cells. Molecules 2019, 24, 3390. https://doi.org/10.3390/molecules24183390
Wang L, Liu M, Yin F, Wang Y, Li X, Wu Y, Ye C, Liu J. Trilobatin, a Novel SGLT1/2 Inhibitor, Selectively Induces the Proliferation of Human Hepatoblastoma Cells. Molecules. 2019; 24(18):3390. https://doi.org/10.3390/molecules24183390
Chicago/Turabian StyleWang, Lujing, Min Liu, Fei Yin, Yuanqiang Wang, Xingan Li, Yucui Wu, Cuilian Ye, and Jianhui Liu. 2019. "Trilobatin, a Novel SGLT1/2 Inhibitor, Selectively Induces the Proliferation of Human Hepatoblastoma Cells" Molecules 24, no. 18: 3390. https://doi.org/10.3390/molecules24183390
APA StyleWang, L., Liu, M., Yin, F., Wang, Y., Li, X., Wu, Y., Ye, C., & Liu, J. (2019). Trilobatin, a Novel SGLT1/2 Inhibitor, Selectively Induces the Proliferation of Human Hepatoblastoma Cells. Molecules, 24(18), 3390. https://doi.org/10.3390/molecules24183390