Synthesis of Isorhamnetin-3-O-Rhamnoside by a Three-Enzyme (Rhamnosyltransferase, Glycine Max Sucrose Synthase, UDP-Rhamnose Synthase) Cascade Using a UDP-Rhamnose Regeneration System
Abstract
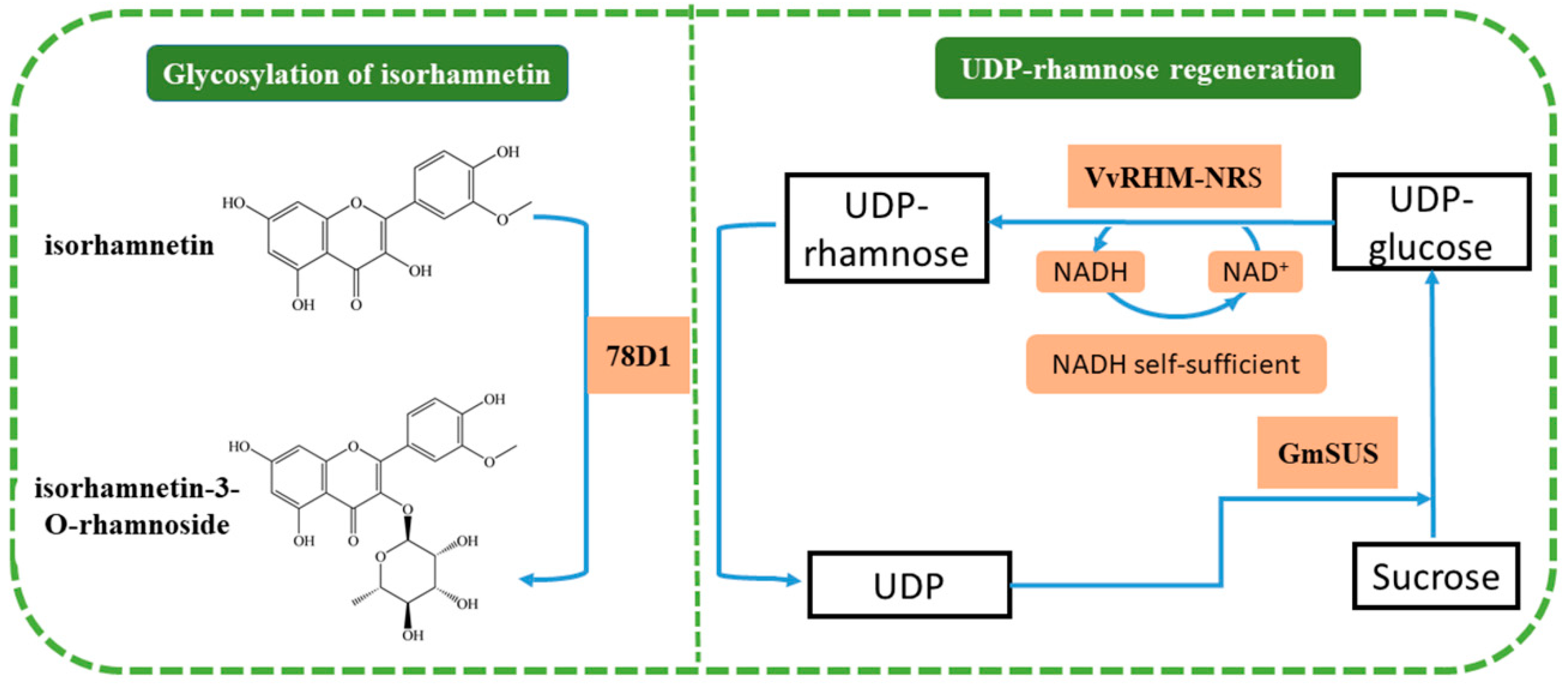
:1. Introduction
2. Results and Discussion
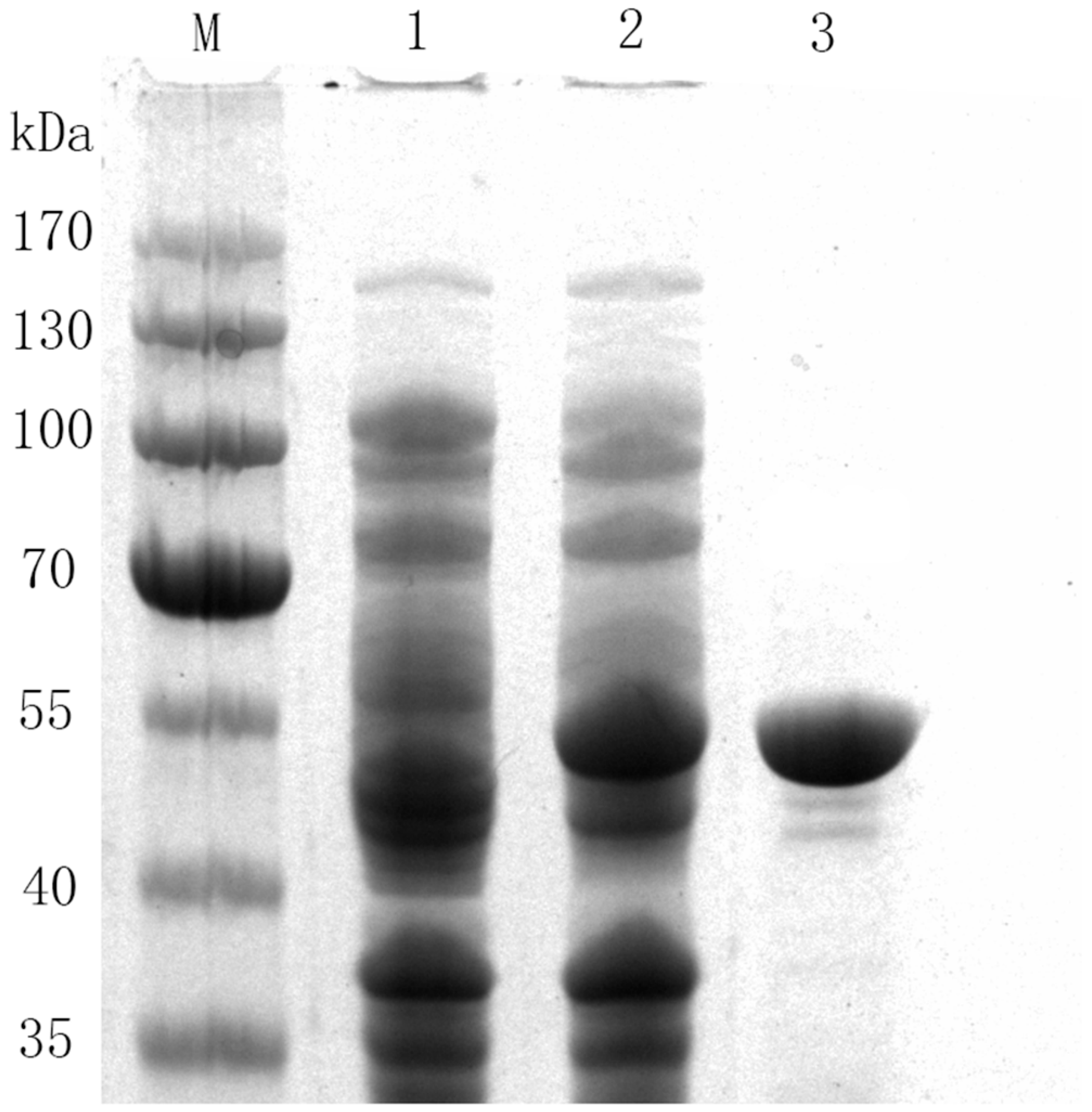
2.1. Characterization and Purification of 78D1
2.2. Optimizing the Ratio among 78D1, GmSUS, and VvRHM-NRS in a One-Pot Synthesis of Isorhamnetin-3-O-Rhamnoside
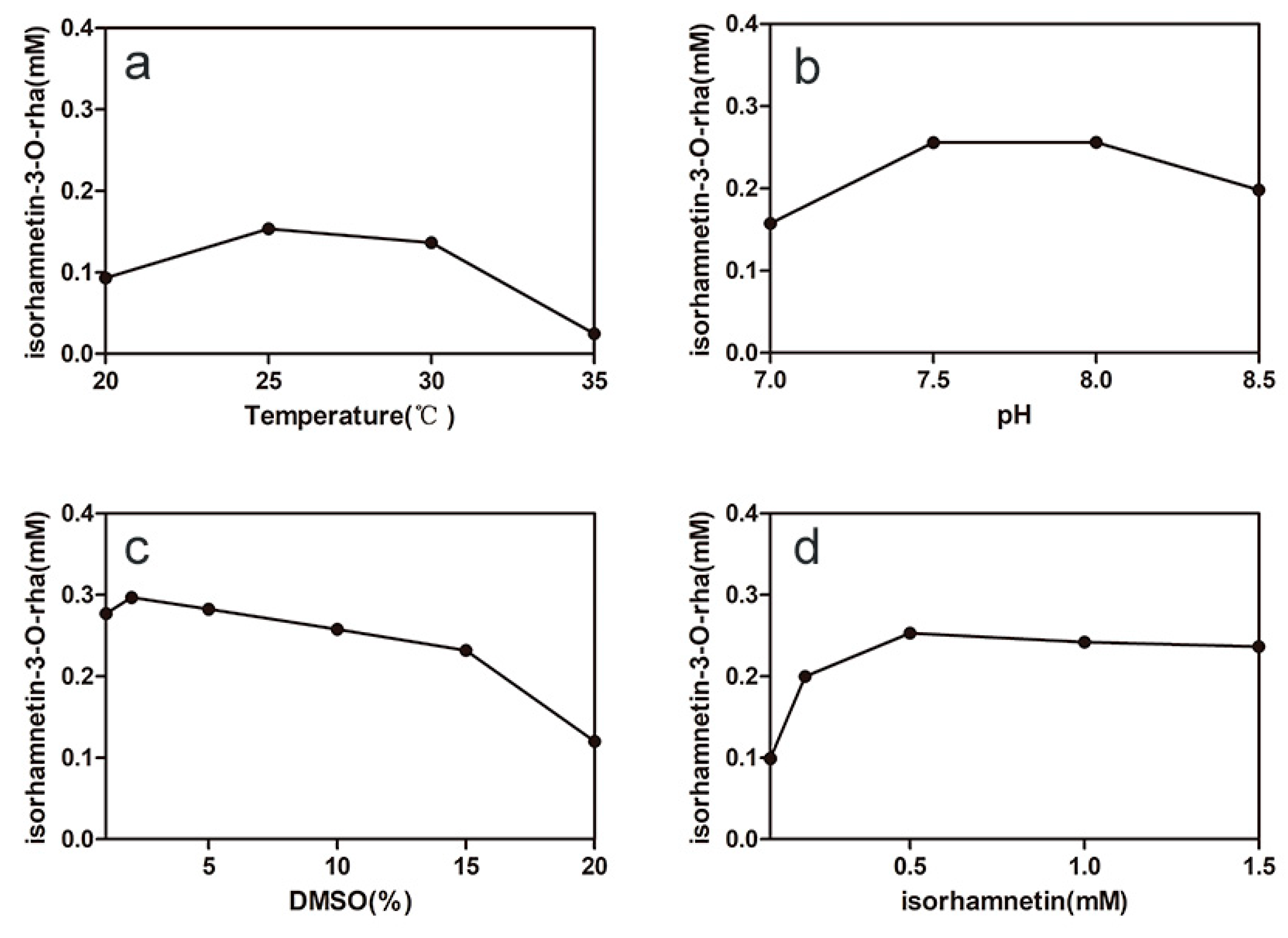
2.3. Optimizing the Conditions of Synergistic Catalysis
2.4. Effects of Uridine Diphosphate (UDP), NAD+, and Sucrose on the One-Pot Synthesis of Isorhamnetin-3-O-rhamnoside
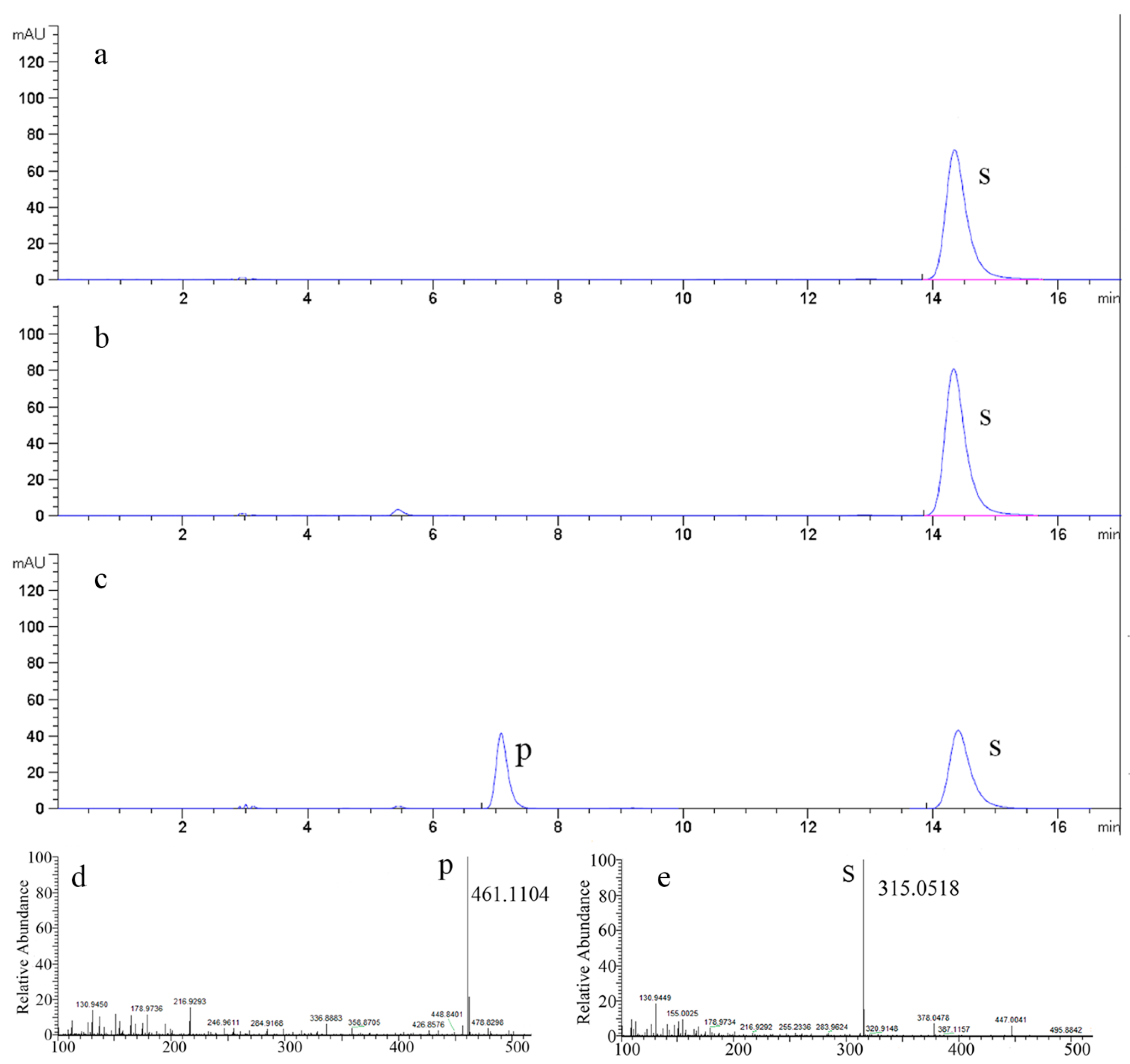
2.5. The Time Courses of Isorhamnetin-3-O-rhamnoside Production and Preparation
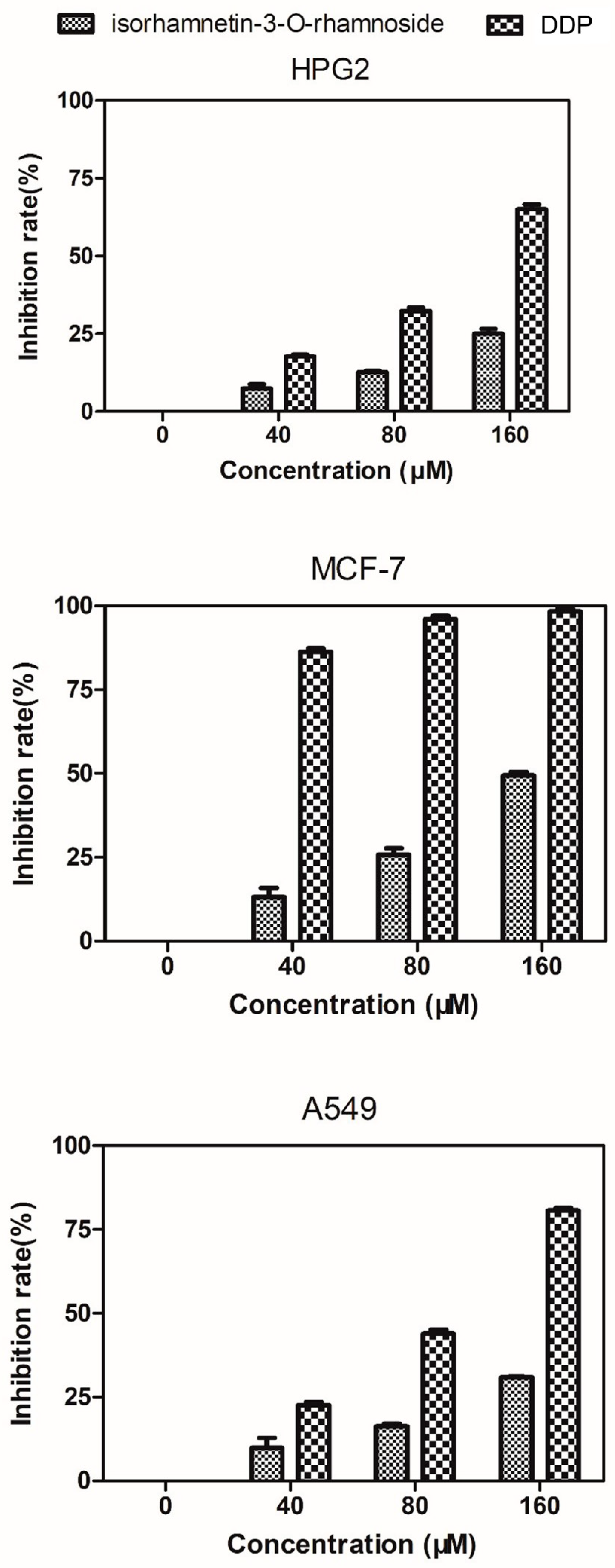
2.6. Evaluation of Cytotoxicity against HepG2, MCF-7, and A549 Cells
3. Materials and Methods
3.1. Strains, Plasmids, Media, and Chemicals
3.2. Plasmid Construction
3.3. Purification of Recombinant Enzymes
3.4. Glycosyltransferase Activity
3.5. Synergistic Catalysis
3.6. Synthesis and Purification of Isorhamnetin-3-O-Rhamnoside
3.7. HPLC, LC/MS Analysis and Structural Identification
3.8. Cell Culture and Cytotoxicity Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Antunes-Ricardo, M.; Gutierrez-Uribe, J.A.; Lopez-Pacheco, F.; Alvarez, M.M.; Serna-Saldivar, S.O. In vivo anti-inflammatory effects of isorhamnetin glycosides isolated from Opuntia ficus-indica (L.) Mill cladodes. Ind. Crop. Prod. 2015, 76, 803–808. [Google Scholar] [CrossRef]
- Kucukboyaci, N.; Akkol, E.K.; Suntar, I.; Calis, I. In vivo Anti-Inflammatory and Antinociceptive Activities of the Extracts and Chemical Constituents of an Endemic Turkish Plant, Salsola grandis. Rec. Nat. Prod. 2016, 10, 369–379. [Google Scholar]
- Park, J.Y.; Kim, S.I.; Lee, H.J.; Kim, S.S.; Kwon, Y.S.; Chun, W. Isorhamnetin-3-O-Glucuronide Suppresses JNK and p38 Activation and Increases Heme-Oxygenase-1 in Lipopolysaccharide-Challenged RAW264.7 Cells. Drug Dev. Res. 2016, 77, 143–151. [Google Scholar] [CrossRef]
- Ku, S.K.; Kim, T.H.; Lee, S.; Kim, S.M.; Bae, J.S. Antithrombotic and profibrinolytic activities of isorhamnetin-3-O-galactoside and hyperoside. Food Chem. Toxicol. 2013, 53, 197–204. [Google Scholar] [CrossRef]
- Boubaker, J. Isorhamnetin 3-O-robinobioside from Nitraria retusa leaves enhance antioxidant and antigenotoxic activity in human chronic myelogenous leukemia cell line K562. BMC Complement. Altern. Med. 2012, 12, 135. [Google Scholar] [CrossRef]
- Chang, L.C.; Tsai, T.R.; Wang, J.J.; Lin, C.N.; Kuo, K.W. The rhamnose moiety of solamargine plays a crucial role in triggering cell death by apoptosis. Biochem. Biophys. Res. Commun. 1998, 242, 21–25. [Google Scholar] [CrossRef]
- Hayder, N.; Bouhlel, I.; Skandrani, I.; Kadri, M.; Steiman, R.; Guiraud, P.; Mariotte, A.; Ghedira, K.; Dijoux-Franca, M.; Chekir-Ghedira, L. In vitro antioxidant and antigenotoxic potentials of myricetin-3-O-galactoside and myricetin-3-O-rhamnoside from Myrtus communis: Modulation of expression of genes involved in cell defence system using cDNA microarray. Toxicol. Vitr. 2008, 22, 567–581. [Google Scholar] [CrossRef]
- Choi, H.J.; Kim, J.H.; Lee, C.H.; Ahn, Y.J.; Song, J.H.; Baek, S.H.; Kwon, D.H. Antiviral activity of quercetin 7-rhamnoside against porcine epidemic diarrhea virus. Antivir. Res. 2009, 81, 77–81. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.J.; Song, J.H.; Park, K.S.; Kwon, D.H. Inhibitory effects of quercetin 3-rhamnoside on influenza A virus replication. Eur. J. Pharm. Sci. 2009, 37, 329–333. [Google Scholar] [CrossRef] [PubMed]
- Atta, E.M.; Hashem, A.I.; Ahmed, A.M.; Elqosyd, S.M.; Jasparse, M.; El-Sharkawc, E.M. Phytochemical studies on Diplotaxis harra growing in Sinai. Eur. J. Chem. 2011, 2, 535–538. [Google Scholar] [CrossRef]
- Osman, S.M.; El-Haddad, A.E.; El-Raey, M.A.; Abd El-Khalik, S.M.; Koheil, M.A.; Wink, M. A New Octadecenoic Acid Derivative from Caesalpinia gilliesii Flowers with Potent Hepatoprotective Activity. Pharmacogn. Mag. 2016, 12, S332–S336. [Google Scholar] [CrossRef] [PubMed]
- Chang, F.R.; Wei, J.L.; Teng, C.M.; Wu, Y.C. Antiplatelet aggregation constituents from Annona purpurea. J. Nat. Prod. 1998, 61, 1457–1461. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Ambrosi, M.; Lopez-Marquez, D.M.; Abad-Garcia, B.; Dapena, E.; Berrueta, L.A.; Gallo, B. Comparative study of phenolic profile of fruit and juice samples of a progeny of ‘Meana’ x ‘Florina’ from an Asturian cider apple breeding program. Eur. Food Res. Technol. 2015, 241, 769–784. [Google Scholar] [CrossRef]
- Osman, S.M.; Abd El-Khalik, S.M.; Abd El-Khalik, A.E.; Wink, M. A new steroidal compound (β-sitosterol-3-O-butyl) isolated from Caesalpinia gilliesii flowers. Int. J. Appl. Res. Nat. Prod. 2015, 8, 14–19. [Google Scholar]
- Kim, B.G.; Su, H.S.; Na, R.J.; Chong, Y.; Ahn, J.H. Biological synthesis of isorhamnetin 3-O-glucoside using engineered glucosyltransferase. J. Mol. Catal. B Enzym. 2010, 63, 194–199. [Google Scholar] [CrossRef]
- Kim, B.G.; Kim, H.J.; Ahn, J.H. Production of bioactive flavonol rhamnosides by expression of plant genes in Escherichia coli. J. Agric. Food Chem. 2012, 60, 11143–11148. [Google Scholar] [CrossRef]
- Lim, E.K.; Ashford, D.A.; Hou, B.K.; Jackson, R.G.; Bowles, D.J. Arabidopsis glycosyltransferases as biocatalysts in fermentation for regioselective synthesis of diverse quercetin glucosides. Biotechnol. Bioeng. 2010, 87, 623–631. [Google Scholar] [CrossRef]
- Kim, H.J.; Kim, B.G.; Ahn, J.H. Regioselective synthesis of flavonoid bisglycosides using Escherichia coli harboring two glycosyltransferases. Appl. Microbiol. Biotechnol. 2013, 97, 5275–5282. [Google Scholar] [CrossRef]
- Lairson, L.L.; Henrissat, B.; Davies, G.J.; Withers, S.G. Glycosyltransferases: Structures, functions, and mechanisms. Annu. Rev. Biochem. 2008, 77, 521. [Google Scholar] [CrossRef]
- Pei, J.; Dong, P.; Wu, T.; Zhao, L.; Fang, X.; Cao, F.; Tang, F.; Yue, Y. Metabolic Engineering of Escherichia coli for Astragalin Biosynthesis. J. Agric. Food. Chem. 2016, 64, 7966–7972. [Google Scholar] [CrossRef]
- Diantini, A.; Subarnas, A.; Lestari, K.; Halimah, E.; Susilawati, Y.; Supriyatna; Julaeha, E.; Achmad, T.H.; Suradji, E.W.; Yamazaki, C.; et al. Kaempferol-3-O-rhamnoside isolated from the leaves of Schima wallichii Korth. inhibits MCF-7 breast cancer cell proliferation through activation of the caspase cascade pathway. Oncol. Lett. 2012, 3, 1069–1072. [Google Scholar] [CrossRef] [PubMed]
- Rho, H.S.; Ghimeray, A.K.; Yoo, D.S.; Ahn, S.M.; Kwon, S.S.; Lee, K.H.; Cho, D.H.; Cho, J.Y. Kaempferol and kaempferol rhamnosides with depigmenting and anti-inflammatory properties. Molecules 2011, 16, 3338. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Ko, C.I.; Ahn, G.; You, S.; Kim, J.S.; Heu, M.S.; Kim, J.; Jee, Y.; Jeon, Y.J. Molecular characteristics and anti-inflammatory activity of the fucoidan extracted from Ecklonia Cava. Carbohydr. Polym. 2012, 89, 599–606. [Google Scholar] [CrossRef] [PubMed]
- Tatsimo, S.J.N.; Tamokou, J.D.D.; Havyarimana, L.; Csupor, D.; Forgo, P.; Hohmann, J.; Kuiate, J.R.; Tane, P. Antimicrobial and antioxidant activity of kaempferol rhamnoside derivatives from Bryophyllum pinnatum. BMC Res. Notes 2012, 5, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Pei, J.J.; Chen, A.; Sun, Q.; Zhao, L.G.; Cao, F.L.; Tang, F. Construction of a novel UDP-rhamnose regeneration system by a two-enzyme reaction system and application in glycosylation of flavonoid. Biochem. Eng. J. 2018, 139, 33–42. [Google Scholar] [CrossRef]
- Rodas, F.R.; Rodriguez, T.O.; Murai, Y.; Iwashina, T.; Sugawara, S.; Suzuki, M.; Nakabayashi, R.; Yonekura-Sakakibara, K.; Saito, K.; Kitajima, J. Linkage mapping, molecular cloning and functional analysis of soybean gene Fg2 encoding flavonol 3-O-glucoside (1 → 6) rhamnosyltransferase. Plant Mol. Biol. 2014, 84, 287–300. [Google Scholar] [CrossRef] [PubMed]
- Casas, M.I.; Falcone-Ferryra, M.L.; Jiang, N.; Mejia Guerra, M.K.; Rodriguez, E.J.; Wilson, T.; Engelmeier, J.; Casati, P.; Grotewold, E. Identification and characterization of maize salmon silks genes involved in insecticidal maysin biosynthesis. Plant Cell 2016, 28, 1297. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.; Messner, B.; Nakajima, J.; Schäffner, A.R.; Saito, K. UGT73C6 and UGT78D1, glycosyltransferases involved in flavonol glycoside biosynthesis in Arabidopsis thaliana. J. Biol. Chem. 2003, 278, 43910–43918. [Google Scholar] [CrossRef]
- Pei, J.; Chen, A.; Zhao, L.; Cao, F.; Ding, G.; Xiao, W. One-pot synthesis of hyperoside by a three-enzyme cascade using a UDP-galactose regeneration system. J. Agric. Food. Chem. 2017, 65, 6042–6048. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are available from the Dr. Jianjun Pei. |




| Entry | 78D1 (μg/mL) | GmSUS (μg/mL) | VvRHM-NRS (μg/mL) | Isorhamnetin-3-O-rhamnoside (mM) |
|---|---|---|---|---|
| 1 | 95 | 8 | 250 | 0.024 |
| 2 | 95 | 16 | 250 | 0.034 |
| 3 | 95 | 32 | 250 | 0.039 |
| 4 | 95 | 88 | 250 | 0.044 |
| 5 | 9.5 | 50 | 250 | 0.015 |
| 6 | 19 | 50 | 250 | 0.025 |
| 7 | 38 | 50 | 250 | 0.033 |
| 8 | 95 | 50 | 250 | 0.042 |
| 9 | 57 | 50 | 125 | 0.014 |
| 10 | 57 | 50 | 188 | 0.017 |
| 11 | 57 | 50 | 250 | 0.041 |
| 12 | 57 | 50 | 375 | 0.059 |
| 13 | 57 | 50 | 500 | 0.064 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, A.; Gu, N.; Pei, J.; Su, E.; Duan, X.; Cao, F.; Zhao, L. Synthesis of Isorhamnetin-3-O-Rhamnoside by a Three-Enzyme (Rhamnosyltransferase, Glycine Max Sucrose Synthase, UDP-Rhamnose Synthase) Cascade Using a UDP-Rhamnose Regeneration System. Molecules 2019, 24, 3042. https://doi.org/10.3390/molecules24173042
Chen A, Gu N, Pei J, Su E, Duan X, Cao F, Zhao L. Synthesis of Isorhamnetin-3-O-Rhamnoside by a Three-Enzyme (Rhamnosyltransferase, Glycine Max Sucrose Synthase, UDP-Rhamnose Synthase) Cascade Using a UDP-Rhamnose Regeneration System. Molecules. 2019; 24(17):3042. https://doi.org/10.3390/molecules24173042
Chicago/Turabian StyleChen, Anna, Na Gu, Jianjun Pei, Erzheng Su, Xuguo Duan, Fuliang Cao, and Linguo Zhao. 2019. "Synthesis of Isorhamnetin-3-O-Rhamnoside by a Three-Enzyme (Rhamnosyltransferase, Glycine Max Sucrose Synthase, UDP-Rhamnose Synthase) Cascade Using a UDP-Rhamnose Regeneration System" Molecules 24, no. 17: 3042. https://doi.org/10.3390/molecules24173042





